Abstract
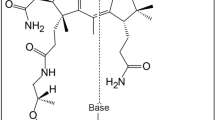
The molybdenum cofactor (Moco) is found in the active site of numerous important enzymes that are critical to biological processes. The bidentate ligand that chelates molybdenum in Moco is the pyranopterin dithiolene (molybdopterin, MPT). However, neither the mechanism of molybdate insertion into MPT nor the structure of Moco prior to its insertion into pyranopterin molybdenum enzymes is known. Here, we report this final maturation step, where adenylated MPT (MPT–AMP) and molybdate are the substrates. X-ray crystallography of the Arabidopsis thaliana Mo-insertase variant Cnx1E S269D D274S identified adenylated Moco (Moco–AMP) as an unexpected intermediate in this reaction sequence. X-ray absorption spectroscopy revealed the first coordination sphere geometry of Moco trapped in the Cnx1E active site. We have used this structural information to deduce a mechanism for molybdate insertion into MPT–AMP. Given their high degree of structural and sequence similarity, we suggest that this mechanism is employed by all eukaryotic Mo-insertases.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The protein structure data that support the findings of this study are publicly available from the Protein Data Bank (https://www.rcsb.org/) with accession code 6Q32. We deposited the protein structure along with the structure factor data file that allows for the re-computing and re-evaluation of the structure. Figures 2 and 3 and Supplementary Figs. 3, 4 and Extended Data Fig. 1 are associated with these raw data. Manuscript datasets are available as Supplementary Data files.
References
Garner, C. D. et al. (eds) RSC Metallobiology Series Vols 1–3 (The Royal Society of Chemistry, 2019); https://doi.org/10.1039/2045-5488
Kirk, M. L. & Stein, B. in Comprehensive Inorganic Chemistry II 2dn edn (eds Reedijk, J. & Poeppelmeier, K.) 263–293 (Elsevier, 2013).
Hille, R., Hall, J. & Basu, P. The mononuclear molybdenum enzymes. Chem. Rev. 114, 3963–4038 (2014).
Stiefel, E. L. The biogeochemistry of molybdenum and tungsten. Met. Ions Biol. Syst. 39, 1–29 (2002).
Schwahn, B. C. et al. Efficacy and safety of cyclic pyranopterin monophosphate substitution in severe molybdenum cofactor deficiency type A: a prospective cohort study. Lancet 386, 1955–1963 (2015).
Mendel, R. R. & Schwarz, G. Molybdenum cofactor biosynthesis in plants and humans. Coord. Chem. Rev. 255, 1145–1158 (2011).
Unkles, S. E. et al. The Aspergillus nidulans cnxABC locus is a single gene encoding two catalytic domains required for synthesis of precursor Z, an intermediate in molybdenum cofactor biosynthesis. J. Biol. Chem. 272, 28381–28390 (1997).
Appleyard, M. et al. The Aspergillus nidulans cnxF gene and its involvement in molybdopterin biosynthesis—molecular characterization and analysis of in vivo generated mutans. J. Biol. Chem. 273, 14869–14876 (1998).
Unkles, S. E., Heck, I. S., Appleyard, M. & Kinghorn, J. R. Eukaryotic molybdopterin synthase—biochemical and molecular studies of Aspergillus nidulans cnxG and cnxH mutants. J. Biol. Chem. 274, 19286–19293 (1999).
Millar, L. et al. Deletion of the CnxE gene encoding the gephyrin-like protein involved in the final stages of molybdenum cofactor biosynthesis in Aspergillus nidulans. Mol. Genet. Genomics 266, 445–453 (2001).
Probst, C. et al. Genetic characterization of the Neurospora crassa molybdenum cofactor biosynthesis. Fungal Genet. Biol. 66, 69–78 (2014).
Mendel, R. R. The molybdenum cofactor. J. Biol. Chem. 288, 13165–13172 (2013).
Weiss, M. C. et al. The physiology and habitat of the last universal common ancestor. Nat. Microbiol 1, 116 (2016).
Hover, B. M., Loksztejn, A., Ribeiro, A. A. & Yokoyama, K. Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. J. Am. Chem. Soc. 135, 7019–7032 (2013).
Hover, B. M., Tonthat, N. K., Schumacher, M. A. & Yokoyama, K. Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis. Proc. Natl Acad. Sci. USA 112, 6347–6352 (2015).
Wuebbens, M. M. & Rajagopalan, K. V. Structural characterization of a molybdopterin precursor. J. Biol. Chem. 268, 13493–13498 (1993).
Teschner, J. et al. A novel role for Arabidopsis mitochondrial ABC transporter ATM3 in molybdenum cofactor biosynthesis. Plant Cell 22, 468–480 (2010).
Wuebbens, M. M. & Rajagopalan, K. V. Mechanistic and mutational studies of Escherichia coli molybdopterin synthase clarify the final step of molybdopterin biosynthesis. J. Biol. Chem. 278, 14523–14532 (2003).
Pitterle, D. M., Johnson, J. L. & Rajagopalan, K. V. In vitro synthesis of molybdopterin from precursor-Z using purified converting factor: role of protein-bound sulfur in formation of the dithiolene. J. Biol. Chem. 268, 13506–13509 (1993).
Leimkuhler, S., Wuebbens, M. M. & Rajagopalan, K. V. The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. Coord. Chem. Rev. 255, 1129–1144 (2011).
Kruse, T. Encyclopedia of Inorganic and Bioinorganic Chemistry https://doi.org/10.1002/9781119951438.eibc2736 (2020).
Arst, H. N., MacDonald, D. W. & Cove, D. J. Molybdate metabolism in Aspergillus nidulans. Mutations affecting nitrate reductase and/or xanthine dehydrogenase. Mol. Gen. Genet. 108, 129–145 (1970).
Joshi, M. S., Johnson, J. L. & Rajagopalan, K. V. Molybdenum cofactor biosynthesis in Escherichia coli mod and mog mutants. J. Bacteriol. 178, 4310–4312 (1996).
Schwarz, G. et al. The molybdenum cofactor biosynthetic protein Cnx1 complements molybdate-repairable mutants, transfers molybdenum to the metal binding pterin, and is associated with the cytoskeleton. Plant Cell 12, 2455–2471 (2000).
Llamas, A., Otte, T., Multhaup, G., Mendel, R. R. & Schwarz, G. The mechanism of nucleotide-assisted molybdenum insertion into molybdopterin—a novel route toward metal cofactor assembly. J. Biol. Chem. 281, 18343–18350 (2006).
Kuper, J., Llamas, A., Hecht, H. J., Mendel, R. R. & Schwarz, G. Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430, 803–806 (2004).
Llamas, A., Mendel, R. R. & Schwarz, N. Synthesis of adenylated molybdopterin—an essential step for molybdenum insertion. J. Biol. Chem. 279, 55241–55246 (2004).
Krausze, J. et al. Dimerization of the plant molybdenum insertase Cnx1E is required for synthesis of the molybdenum cofactor. Biochem. J. 474, 163–178 (2017).
Krausze, J. et al. The functional principle of eukaryotic molybdenum insertases. Biochem. J. 475, 1739–1753 (2018).
Hercher, T. W. et al. Insights into the Cnx1E catalyzed MPT-AMP hydrolysis. Biosci. Rep. 40, BSR20191806 (2020).
Heck, I. S. et al. Mutational analysis of the gephyrin-related molybdenum cofactor biosyndietic gene cnxE from the lower eukaryote Aspergillus nidulans. Genetics 161, 623–632 (2002).
Kirk, M. L. in Molybdenum and Tungsten Enzymes: Spectroscopic and Theoretical Investigations RSC Metallobiology Series No. 7 (eds Hille, R. et al.) 13–67 (The Royal Society of Chemistry, 2016).
Schwarz, G., Boxer, D. H. & Mendel, R. R. in Chemistry and Biology of Pteridines and Folates (eds Pfleiderer, W. & Rokos, H.) 697–702 (Blackwell Science, 1997).
Schwarz, G. & Mendel, R. R. Molybdenum cofactor biosynthesis and molybdoenzymes. Annu. Rev. Plant Biol. 57, 623–647 (2006).
Rothery, R. A., Stein, B., Solomonson, M., Kirk, M. L. & Weiner, J. H. Pyranopterin conformation defines the function of molybdenum and tungsten enzymes. Proc. Natl Acad. Sci. USA 109, 14773–14778 (2012).
Xiang, S., Nichols, J., Rajagopalan, K. V. & Schindelin, H. The crystal structure of Escherichia coli MoeA and its relationship to the multifunctional protein gephyrin. Structure 9, 299–310 (2001).
Kutzler, F. W. et al. Single-crystal polarized X-ray absorption spectroscopy. Observation and theory for (MoO2S2)2−. J. Am. Chem. Soc. 103, 6083–6088 (1981).
George, G. N. et al. Structure of the active site of sulfite oxidase. X-ray absorption spectroscopy of the molybdenum (iv), molybdenum (v) and molybdenum (vi) oxidation states. Biochemistry 28, 5075–5080 (1989).
Partyka, D. V. & Holm, R. H. Oxygen/sulfur substitution reactions of tetraoxometalates effected by electrophilic carbon and silicon reagents. Inorg. Chem. 43, 8609–8616 (2004).
Wang, J. J. & Holm, R. H. Silylation, sulfidation and benzene-1,2-dithiolate complexation reactions of oxo- and oxosulfidomolybdates(vi) and -tungstates(vi). Inorg. Chem. 46, 11156–11164 (2007).
Lim, B. S., Willer, M. W., Miao, M. M. & Holm, R. H. Monodithiolene molybdenum(v,vi) complexes: a structural analogue of the oxidized active site of the sulfite oxidase enzyme family. J. Am. Chem. Soc. 123, 8343–8349 (2001).
Harris, H. H., George, G. N. & Rajagopalan, K. V. High-resolution EXAFS of the active site of human sulfite oxidase: comparison with density functional theory and X-ray crystallographic results. Inorg. Chem. 45, 493–495 (2006).
Liu, X. D., Cheng, J., Sprik, M. & Lu, X. C. Solution structures and acidity constants of molybdic acid. J. Phys. Chem. Lett. 4, 2926–2930 (2013).
George, G. N., Garrett, R. M., Prince, R. C. & Rajagopalan, K. V. The molybdenum site of sulfite oxidase: a comparison of wild-type and the cysteine 207 to serine mutant using X-ray absorption spectroscopy. J. Am. Chem. Soc. 118, 8588–8592 (1996).
Minubayeva, Z. & Seward, T. M. Molybdic acid ionisation under hydrothermal conditions to 300 °C. Geochim. Cosmochim. Acta 74, 4365–4374 (2010).
Schwarz, G., Boxer, D. H. & Mendel, R. R. Molybdenum cofactor biosynthesis—the plant protein Cnx1 binds molybdopterin with high affinity. J. Biol. Chem. 272, 26811–26814 (1997).
Fischer, K. et al. Function and structure of the molybdenum cofactor carrier protein from Chlamydomonas reinhardtii. J. Biol. Chem. 281, 30186–30194 (2006).
Ringel, P. et al. Biochemical characterization of molybdenum cofactor-free nitrate reductase from Neurospora crassa. J. Biol. Chem. 288, 14657–14671 (2013).
Kruse, T. et al. Identification and biochemical characterization of molybdenum cofactor-binding proteins from Arabidopsis thaliana. J. Biol. Chem. 285, 6623–6635 (2010).
Stallmeyer, B., Nerlich, A., Schiemann, J., Brinkmann, H. & Mendel, R. R. Molybdenum cofactor biosynthesis: the Arabidopsis thaliana cDNA cnx1 encodes a multifunctional 2-domain protein homologous to a mammalian neuroprotein, the insect protein Cinnamon and three Escherichia coli proteins. Plant J. 8, 751–762 (1995).
Leslie, A. G. W. & Powell, H. R. in Evolving Methods for Macromolecular Crystallography (eds Read, R. J. & Sussman, J. L.) 41–51 (Springer, 2007).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
STARANISO (Global Phasing Ltd, 2017).
BUSTER v. 2.10.3 (Global Phasing Ltd, 2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Struct. Biol. 67, 235–242 (2011).
The PyMOL Molecular Graphics System v. 1.5.0.4 (Schrödinger, 2020).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Ravel, B. & Newville, M. ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J. Synchrotron Radiat. 12, 537–541 (2005).
Frisch. M. J. et al. Gaussian 16, revision C.01 (Gaussian, Inc., 2016).
Acknowledgements
We thank A. Calean (TU Braunschweig) for excellent support with inductively coupled plasma mass spectrometry analysis. M.L.K. gratefully acknowledges support of this research by the National Institutes of Health (grant no. GM-057378). M.L.K. and J.Y. thank the UNM Center for Advanced Research Computing, supported in part by the National Science Foundation, for providing the computing resources used in this work. Work at Caltech was supported by the National Institutes of Health (NIH) grant no. GM045162. We acknowledge the Gordon and Betty Moore Foundation and the Beckman Institute at Caltech for their support of the Molecular Observatory at Caltech and the staff at beamlines 12-2 and 7-3. M.L.K. and J.Y. acknowledge the Stanford Synchrotron Radiation Lightsource, which is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. R.R.M. and T.K. acknowledge the support of this research by the Deutsche Forschungsgemeinschaft (GRK 2223/1). Correspondence and requests for materials should be addressed to M.L.K. and T.K.
Author information
Authors and Affiliations
Contributions
C.P. carried out acquisition, analysis and interpretation of data. J.Y. carried out acquisition, analysis and interpretation of XAS data and computed the reaction coordinate. J.K. analysed and interpreted the data. T.W.H. carried out acquisition, analysis and interpretation of data. C.P.R. synthesized and characterized the trioxo- and dioxo-molybdenum model compounds (2, 3 and 4) and analysed spectroscopic data. T.S. carried out acquisition, analysis and interpretation of data. K.K. assisted with the collection of XAS data. L.J.G. assisted with the collection of XAS data. D.C.R. and R.R.M. revised the manuscript. M.L.K. conceived the idea, performed design analysis and interpretation of the data, and drafted the manuscript. T.K. conceived the idea and design, analysed and interpreted the data, and drafted the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemistry thanks Partha Basu, Carola Schulzke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The proposed mechanism of molybdate insertion into the MPT dithiolene moiety.
a, Ground state of Cnx1E with a Mg2+-water complex bound. Mg2+ constitutes the cofactor for the Cnx1E catalyzed reaction25,30. b, Conformation of MPT-AMP bound in the active site of Cnx1G26. c, First state of the catalysis with the substrate molybdate bound to the entry site. The presence of molybdate in the entry site induces an unfavorable (tense) backbone conformation in Cnx1E29. d, The conformation of MPT-AMP found in Cnx1G is not compatible with the Cnx1E active site28. Hence, the MPT-AMP pterin moiety must re-orient upon transfer from Cnx1G to Cnx1E, mostly involving two bond rotations. e, Second state of the catalytic cycle with molybdate bound to the entry site and active site bound MPT-AMP. The orientation of the pterin moiety was derived from active site bound Moco-AMP identified in this work. f, The transfer of molybdate from the entry to the insertion site is assisted by the relaxation of the tense backbone conformation29. According to our model of molybdate insertion, one molybdate oxygen ligand is two-fold protonated by the MPT dithiolene function and released as water. Simultaneously, the sulfur to molybdenum bonds are formed: This mechanism begins with a proton transfer from the diacid form of the MPT dithiolene side chain to a molybdate oxygen. A second, less activated protonation on the same oxygen atom would then yield a [(MPT)MoO3(OH2)]2- transition state. This [(MPT)MoO3(OH2)]2-species is expected to possess a highly labile aqua ligand, which would subsequently dissociate from Mo along the reaction coordinate to yield a more stable trioxo [(MPT)MoO3]2- intermediate. A final protonation of basic [(MPT)MoO3]2- by a suitable Cnx1E active site residue (most likely residue Arg369) yields our XAS derived [(MPT)MoO2(OH)]1- structure for Cnx1E Moco-AMP (for details see Fig. 5 and supplementary Fig. 11). The protonated oxygen atom is marked by an asterisk. g, The Cnx1E active site with bound Moco-AMP.
Supplementary information
Supplementary Information
Supplementary Figs 1–11 and Tables 1–5.
Supplementary Data 1
Primary data (dioxo (3)) Fig. 4a.
Supplementary Data 2
Primary data (insertase pH 6) Fig. 4a.
Supplementary Data 3
Primary data (insertase pH 8) Fig. 4a.
Supplementary Data 4
Primary data (molybdate) Fig. 4a.
Supplementary Data 5
Primary data (trioxo (2)) Fig. 4a.
Supplementary Data 6
Evaluated data Fig. 4b.
Supplementary Data 7
Primary data Fig.4c.
Supplementary Data 8
Primary data Fig. 4e.
Supplementary Data 9
Primary data Fig. 4f.
Supplementary Data 10
Primary data Supplementary Fig. 5a.
Supplementary Data 11
Primary data Supplementary Fig. 5b.
Supplementary Data 12
Primary data Supplementary Fig. 5c.
Supplementary Data 13
Primary HPLC data and evaluated data Supplementary Fig. 1a, replicate 1.
Supplementary Data 14
Calibration report replicate 1, Supplementary Fig. 1a.
Supplementary Data 15
Primary HPLC data and evaluated data Supplementary Fig. 1a, replicates 2 and 3.
Supplementary Data 16
Calibration report replicates 2 and 3, Supplementary Fig. 1a.
Supplementary Data 17
Primary ICP-MS Data and evaluated Data Suppl. Fig.1 A, Replicates 1–3.
Rights and permissions
About this article
Cite this article
Probst, C., Yang, J., Krausze, J. et al. Mechanism of molybdate insertion into pterin-based molybdenum cofactors. Nat. Chem. 13, 758–765 (2021). https://doi.org/10.1038/s41557-021-00714-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-021-00714-1