Abstract
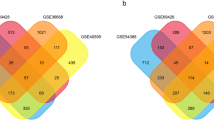
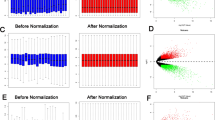
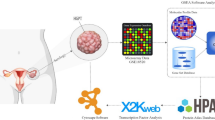
Ovarian cancer (OVC) is often diagnosed at the advanced stage resulting in a poor overall outcome for the patient. The disease mechanisms, prognosis, and treatment require imperative elucidation. A rank-based module-centric framework was proposed to analyze the key modules related to the development, prognosis, and treatment of OVC. The ovarian cancer cell line microarray dataset GSE43765 from the Gene Expression Omnibus database was used to construct the reference modules by weighted gene correlation network analysis. Twenty-three reference modules were tested for stability and functionally annotated. Furthermore, to demonstrate the utility of reference modules, two more OVC datasets were collected, and their gene expression profiles were projected to the reference modules to generate a module-level expression. An epithelial-mesenchymal transition module was activated in OVC compared to the normal epithelium, and a pluripotency module was activated in ovarian cancer stroma compared to ovarian cancer epithelium. Seven differentially expressed modules were identified in OVC compared to the normal ovarian epithelium, with five up-regulated, and two down-regulated. One module was identified to be predictive of patient overall survival. Four modules were enriched with SNP signals. Based on differentially expressed modules and hub genes, five candidate drugs were screened. The hub genes of those modules merit further investigation. We firstly propose the reference module-based analysis of OVC. The utility of the analysis framework can be extended to transcriptome data of other kinds of diseases.








Similar content being viewed by others
Data Availability
The datasets analyzed in the study are available at the public database NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/). The code used to generate the main result has been made available on GitHub (https://github.com/mrliuw/RefNet).
References
Allan D, Tieu A, Lalu M, Burger D (2020) Mesenchymal stromal cell-derived extracellular vesicles for regenerative therapy and immune modulation: progress and challenges toward clinical application. Stem Cells Transl Med 9:39–46
Barabási A-L, Gulbahce N, Loscalzo J (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12:56–68
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M (2012) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41:D991–D995
Bartlett TE, Müller S, Diaz A (2017) Single-cell co-expression subnetwork analysis. Sci Rep 7:15066
Chen HT, Liu H, Mao MJ, Tan Y, Mo XQ, Meng XJ, Cao MT, Zhong CY, Liu Y, Shan H, Jiang GM (2019) Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol Cancer 18:101
Christoforou N, Chellappan M, Adler AF, Kirkton RD, Wu T, Addis RC, Bursac N, Leong KW (2013) Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS ONE 8:e63577
Chung VY, Tan TZ, Ye J, Huang RL, Lai HC, Kappei D, Wollmann H, Guccione E, Huang RY (2019) The role of GRHL2 and epigenetic remodeling in epithelial-mesenchymal plasticity in ovarian cancer cells. Commun Biol 2:272
Cortes J, Schoffski P, Littlefield BA (2018) Multiple modes of action of eribulin mesylate: emerging data and clinical implications. Cancer Treat Rev 70:190–198
Cox DR (1972) Regression models and life-tables. J R Stat Soc B 34:187–220
D’Haeseleer P, Liang S, Somogyi R (2000) Genetic network inference: from co-expression clustering to reverse engineering. Bioinformatics 16:707–726
Dezso Z, Oestreicher J, Weaver A, Santiago S, Agoulnik S, Chow J, Oda Y, Funahashi Y (2014) Gene expression profiling reveals epithelial mesenchymal transition (EMT) genes can selectively differentiate eribulin sensitive breast cancer cells. PLoS ONE 9:e106131
Fiscon G, Conte F, Licursi V, Nasi S, Paci P (2018) Computational identification of specific genes for glioblastoma stem-like cells identity. Sci Rep 8:7769
Fiscon G, Paci P (2021) SAveRUNNER: an R-based tool for drug repurposing. BMC Bioinformatics 22:150
Gu KL, Zhang Q, Yan Y, Li TT, Duan FF, Hao J, Wang XW, Shi M, Wu DR, Guo WT, Wang Y (2016) Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res 26:350–366
Harr B, Schlötterer C (2006) Comparison of algorithms for the analysis of Affymetrix microarray data as evaluated by co-expression of genes in known operons. Nucleic Acids Res 34:e8–e8
Haston KM, Tung JY, Reijo Pera RA (2009) Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS ONE 4:e5654
Hattori K, Heissig B, Wu Y, Dias S, Tejada R, Ferris B, Hicklin DJ, Zhu Z, Bohlen P, Witte L, Hendrikx J, Hackett NR, Crystal RG, Moore MA, Werb Z, Lyden D, Rafii S (2002) Placental growth factor reconstitutes hematopoiesis by recruiting VEGFR1(+) stem cells from bone-marrow microenvironment. Nat Med 8:841–849
Heinäniemi M, Nykter M, Kramer R, Wienecke-Baldacchino A, Sinkkonen L, Zhou JX, Kreisberg R, Kauffman SA, Huang S, Shmulevich I (2013) Gene-pair expression signatures reveal lineage control. Nat Methods 10:577–583
Hensley ML, Kravetz S, Jia X, Iasonos A, Tew W, Pereira L, Sabbatini P, Whalen C, Aghajanian CA, Zarwan C, Berlin S (2012) Eribulin mesylate (halichondrin B analog E7389) in platinum-resistant and platinum-sensitive ovarian cancer: a 2-cohort, phase 2 study. Cancer 118:2403–2410
Kampan NC, Madondo MT, McNally OM, Quinn M, Plebanski M (2015) Paclitaxel and its evolving role in the management of ovarian cancer. Biomed Res Int 2015:413076
Kurata T, Fushida S, Kinoshita J, Oyama K, Yamaguchi T, Okazaki M, Miyashita T, Tajima H, Ninomiya I, Ohta T (2018) Low-dose eribulin mesylate exerts antitumor effects in gastric cancer by inhibiting fibrosis via the suppression of epithelial-mesenchymal transition and acts synergistically with 5-fluorouracil. Cancer Manag Res 10:2729–2742
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J-P, Subramanian A, Ross KN (2006) The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313:1929–1935
Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1:54
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559
Langfelder P, Luo R, Oldham MC, Horvath S (2011) Is my network module preserved and reproducible? PLoS Comput Biol 7:e1001057
Le YS, Kim TE, Kim BK, Park YG, Kim GM, Jee SB, Ryu KS, Kim IK, Kim JW (2002) Alterations of HLA class I and class II antigen expressions in borderline, invasive and metastatic ovarian cancers. Exp Mol Med 34:18–26
Leutenegger M, Bruckner R, Spalinger MR, Lang S, Rogler G, Scharl M (2018) Eribulin does not prevent epithelial-to-mesenchymal transition in ht-29 intestinal epithelial cells. Inflamm Intest Dis 2:211–218
Li M, Wang Q, Lin W, Wang B (2009) Regulation of ovarian cancer cell adhesion and invasion by chloride channels. Int J Gynecol Cancer 19:526–530
Licursi V, Conte F, Fiscon G, Paci P (2019) MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinformatics 20:545
Lim LS, Loh YH, Zhang W, Li Y, Chen X, Wang Y, Bakre M, Ng HH, Stanton LW (2007) Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol Biol Cell 18:1348–1358
Liu W, Li L, Li W (2014) Gene co-expression analysis identifies common modules related to prognosis and drug resistance in cancer cell lines. Int J Cancer 135:2795–2803
Liu W, Li L, Ye H, Tao H, He H (2018a) Role of COL6A3 in colorectal cancer. Oncol Rep 39:2527–2536
Liu W, Tu W, Li L, Liu Y, Wang S, Li L, Tao H, He H (2018b) Revisiting connectivity map from a gene co-expression network analysis. Exp Ther Med 16:493–500
MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, Pendlington ZM, Welter D, Burdett T, Hindorff L, Flicek P, Cunningham F, Parkinson H (2017) The new NHGRI-EBI catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45:D896–D901
Matsuoka H, Tsurutani J, Tanizaki J, Iwasa T, Komoike Y, Koyama A, Nakagawa K (2013) Regression of brain metastases from breast cancer with eribulin: a case report. BMC Res Notes 6:541
Miller HE, Bishop AJR (2021) Correlation AnalyzeR: functional predictions from gene co-expression correlations. BMC Bioinformatics 22:206
Mousavian Z, Nowzari-Dalini A, Stam RW, Rahmatallah Y, Masoudi-Nejad A (2017) Network-based expression analysis reveals key genes related to glucocorticoid resistance in infant acute lymphoblastic leukemia. Cell Oncol (dordr) 40:33–45
Niemira M, Collin F, Szalkowska A, Bielska A, Chwialkowska K, Reszec J, Niklinski J, Kwasniewski M, Kretowski A (2019) Molecular signature of subtypes of non-small-cell lung cancer by large-scale transcriptional profiling: identification of key modules and genes by weighted gene co-expression network analysis (WGCNA). Cancers 12(1):37
Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH (2008) Functional organization of the transcriptome in human brain. Nat Neurosci 11:1271–1282
Paci P, Colombo T, Fiscon G, Gurtner A, Pavesi G, Farina L (2017) SWIM: a computational tool to unveiling crucial nodes in complex biological networks. Sci Rep 7:44797
Paci P, Fiscon G, Conte F, Wang R-S, Farina L, Loscalzo J (2021) Gene co-expression in the interactome: moving from correlation toward causation via an integrated approach to disease module discovery. Npj Syst Biol Appl 7:3
Pederson PEJ, Mooberry SL (2020) Abstract P6-07-02: evaluating the effects of eribulin and paclitaxel on exosome formation and release from triple negative breast cancer cells. Cancer Res. https://doi.org/10.1158/1538-7445
Popovic J, Stanisavljevic D, Schwirtlich M, Klajn A, Marjanovic J, Stevanovic M (2014) Expression analysis of SOX14 during retinoic acid induced neural differentiation of embryonal carcinoma cells and assessment of the effect of its ectopic expression on SOXB members in HeLa cells. PLoS ONE 9:e91852
Pozzi A, Capdevila JH (2008) PPARalpha ligands as antitumorigenic and antiangiogenic agents. PPAR Res 2008:906542
Qiu X, Cheng SH, Xu F, Yin JW, Wang LY, Zhang XY (2020) Weighted gene co-expression network analysis identified MYL9 and CNN1 are associated with recurrence in colorectal cancer. J Cancer 11:2348–2359
Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J (2016) g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 44:W83-89
Samuel P, Mulcahy LA, Furlong F, McCarthy HO, Brooks SA, Fabbri M, Pink RC, Carter DRF (2018) Cisplatin induces the release of extracellular vesicles from ovarian cancer cells that can induce invasiveness and drug resistance in bystander cells. Philos Trans R Soc Lond B Biol Sci 373:20170065
Shen H, Fridley BL, Song H, Lawrenson K, Cunningham JM, Ramus SJ, Cicek MS, Tyrer J, Stram D, Larson MC, Kobel M, Consortium P, Ziogas A, Zheng W, Yang HP, Wu AH, Wozniak EL, Woo YL, Winterhoff B, Wik E, Whittemore AS, Wentzensen N, Weber RP, Vitonis AF, Vincent D, Vierkant RA, Vergote I, Van Den Berg D, Van Altena AM, Tworoger SS, Thompson PJ, Tessier DC, Terry KL, Teo SH, Templeman C, Stram DO, Southey MC, Sieh W, Siddiqui N, Shvetsov YB, Shu XO, Shridhar V, Wang-Gohrke S, Severi G, Schwaab I, Salvesen HB, Rzepecka IK, Runnebaum IB, Rossing MA, Rodriguez-Rodriguez L, Risch HA, Renner SP, Poole EM, Pike MC, Phelan CM, Pelttari LM, Pejovic T, Paul J, Orlow I, Omar SZ, Olson SH, Odunsi K, Nickels S, Nevanlinna H, Ness RB, Narod SA, Nakanishi T, Moysich KB, Monteiro AN, Moes-Sosnowska J, Modugno F, Menon U, McLaughlin JR, McGuire V, Matsuo K, Adenan NA, Massuger LF, Lurie G, Lundvall L, Lubinski J, Lissowska J, Levine DA, Leminen A, Lee AW, Le ND, Lambrechts S, Lambrechts D, Kupryjanczyk J, Krakstad C, Konecny GE, Kjaer SK, Kiemeney LA, Kelemen LE, Keeney GL, Karlan BY, Karevan R, Kalli KR, Kajiyama H, Ji BT, Jensen A, Jakubowska A, Iversen E, Hosono S, Hogdall CK, Hogdall E, Hoatlin M, Hillemanns P, Heitz F, Hein R, Harter P, Halle MK, Hall P, Gronwald J, Gore M, Goodman MT, Giles GG, Gentry-Maharaj A, Garcia-Closas M, Flanagan JM, Fasching PA, Ekici AB, Edwards R, Eccles D, Easton DF, Durst M, du Bois A, Dork T, Doherty JA, Despierre E, Dansonka-Mieszkowska A, Cybulski C, Cramer DW, Cook LS, Chen X, Charbonneau B, Chang-Claude J, Campbell I, Butzow R, Bunker CH, Brueggmann D, Brown R, Brooks-Wilson A, Brinton LA, Bogdanova N, Block MS, Benjamin E, Beesley J, Beckmann MW, Bandera EV, Baglietto L, Bacot F, Armasu SM, Antonenkova N, Anton-Culver H, Aben KK, Liang D, Wu X, Lu K, Hildebrandt MA, Australian Ovarian Cancer Study G, Australian Cancer S, Schildkraut JM, Sellers TA, Huntsman D, Berchuck A, Chenevix-Trench G, Gayther SA, Pharoah PD, Laird PW, Goode EL, Pearce CL (2013) Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Commun 4:1628
Silverman EK, Schmidt H (2020) Molecular networks in network medicine: development and applications. Wiley Interdiscip Rev Syst Biol Med 12:e1489
Skinner J, Kotliarov Y, Varma S, Mine KL, Yambartsev A, Simon R, Huyen Y, Morgun A (2011) Construct and compare gene coexpression networks with DAPfinder and DAPview. BMC Bioinformatics 12:286
Sun Q, Yogosawa S, Iizumi Y, Sakai T, Sowa Y (2015) The alkaloid emetine sensitizes ovarian carcinoma cells to cisplatin through downregulation of bcl-xL. Int J Oncol 46:389–394
Swaffar DS, Ang CY, Desai PB, Rosenthal GA (1994) Inhibition of the growth of human pancreatic cancer cells by the arginine antimetabolite L-canavanine. Cancer Res 54:6045–6048
Team RC (2016) R: a language and environment for statistical computing. Vienna, Austria
Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, Traficante N, Fereday S, Hung JA, Chiew YE, Haviv I, Gertig D, DeFazio A, Bowtell DD, Australian Ovarian Cancer Study G (2008) Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 14:5198–5208
Vincent KM, Postovit L-M (2017) A pan-cancer analysis of secreted Frizzled-related proteins: re-examining their proposed tumour suppressive function. Sci Rep 7:42719
Wang H, Sun Q, Zhao W, Qi L, Gu Y, Li P, Zhang M, Li Y, Liu S-L, Guo Z (2014) Individual-level analysis of differential expression of genes and pathways for personalized medicine. Bioinformatics 31:62–68
Wang X, Chai Z, Li Y, Long F, Hao Y, Pan G, Liu M, Li B (2020) Identification of potential biomarkers for Anti-PD-1 therapy in melanoma by weighted correlation network analysis. Genes 11:435
Yuan Y, Chen J, Wang J, Xu M, Zhang Y, Sun P, Liang L (2020) Identification hub genes in colorectal cancer by integrating weighted gene co-expression network analysis and clinical validation in vivo and vitro. Front Oncol 10:638
Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. https://doi.org/10.2202/1544-6115.1128
Zhang H, Du Y, Wang Z, Lou R, Wu J, Feng J (2019) Integrated analysis of oncogenic networks in colorectal cancer identifies GUCA2A as a molecular marker. Biochem Res Int 2019:6469420
Funding
This work was supported by the Department of Science and Technology of Fuzhou City under Grant Nos. 2018-S-105-4 and 2017-S-129-3; the Construction Project for Clinical Medicine Center in Fujian Province under Grant No. 201610192; and the Natural Science Foundation of Fujian Province under Grant No. 2019D004.
Author information
Authors and Affiliations
Contributions
ZL, XL, and PL designed the study; JY, WL, and SL collected and analyzed the data; XL and PL analyzed the results, plotted the figures, and drafted the manuscript; All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lai, X., Lin, P., Ye, J. et al. Reference Module-Based Analysis of Ovarian Cancer Transcriptome Identifies Important Modules and Potential Drugs. Biochem Genet 60, 433–451 (2022). https://doi.org/10.1007/s10528-021-10101-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10101-7