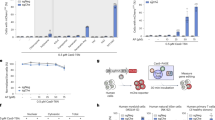
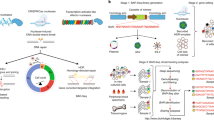
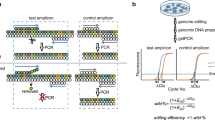
Abstract
Deaminase fused-Cas9 base editing technologies have enabled precise single-nucleotide genomic editing without the need for the introduction of damaging double-stranded breaks and inefficient homology-directed repair. However, current methods to isolate base-edited cell populations are ineffective, especially when utilized with human pluripotent stem cells, a cell type resistant to genome modification. Here, we outline a series of methods that employ transient reporters of editing enrichment (TREE) to facilitate the highly efficient single-base editing of human cells at precise genomic loci. Briefly, these transient reporters of editing enrichment based methods employ a transient episomal fluorescent reporter that allows for the real-time, flow-cytometry-based enrichment of cells that have had single nucleotide changes at precise genomic locations. This protocol details how these approaches can enable the rapid (~3–4 weeks) and efficient (clonal editing efficiencies >80%) generation of biallelic or multiplexed edited isogenic hPSC lines using adenosine and cytosine base editors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Hockemeyer, D. & Jaenisch, R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell 18, 573–586 (2016).
Buecker, C. et al. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell 6, 535–546 (2010).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Joung, J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
De Masi, C., Spitalieri, P., Murdocca, M., Novelli, G. & Sangiuolo, F. Application of CRISPR/Cas9 to human-induced pluripotent stem cells: from gene editing to drug discovery. Hum. Genomics 14, 25 (2020).
Chang, C.-Y., Ting, H.-C., Su, H.-L. & Jeng, J.-R. Combining induced pluripotent stem cells and genome editing technologies for clinical applications. Cell Transplant. 27, 379–392 (2018).
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
Yu, C. et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16, 142–147 (2015).
Takayama, K. et al. Highly efficient biallelic genome editing of human ES/iPS cells using a CRISPR/Cas9 or TALEN system. Nucleic Acids Res. 45, 5198–5207 (2017).
Arias-Fuenzalida, J. et al. FACS-assisted CRISPR-Cas9 genome editing facilitates Parkinson’s disease modeling. Stem Cell Rep. 9, 1423–1431 (2017).
Steyer, B. et al. Scarless genome editing of human pluripotent stem cells via transient puromycin selection. Stem Cell Rep. 10, 642–654 (2018).
Liang, G. & Zhang, Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell 13, 149–159 (2013).
Omole, A. E. & Fakoya, A. O. J. Ten years of progress and promise of induced pluripotent stem cells: historical origins, characteristics, mechanisms, limitations, and potential applications. PeerJ 6, e4370 (2018).
Martin, R. M. et al. Highly efficient and marker-free genome editing of human pluripotent stem cells by CRISPR-Cas9 RNP and AAV6 donor-mediated homologous recombination. Cell Stem Cell 24, 821–828.e5 (2019).
Riesenberg, S. & Maricic, T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat. Commun. 9, 2164 (2018).
Ihry, R. J. et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018).
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. & Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930 (2018).
Li, X.-L. et al. Highly efficient genome editing via CRISPR-Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 46, 10195–10215 (2018).
Merkle, F. T. et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 545, 229–233 (2017).
Chapman, J. R., Taylor, M. R. G. & Boulton, S. J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 (2012).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Yang, B., Yang, L. & Chen, J. Development and application of base editors. CRISPR J. 2, 91–104 (2019).
Eid, A., Alshareef, S. & Mahfouz, M. M. CRISPR base editors: genome editing without double-stranded breaks. Biochem. J. 475, 1955–1964 (2018).
Brookhouser, N. et al. BIG-TREE: base-edited isogenic hPSC line generation using a transient reporter for editing enrichment. Stem Cell Rep. 14, 184–191 (2020).
Raman, S., Brookhouser, N. & Brafman, D. A. Using human induced pluripotent stem cells (hiPSCs) to investigate the mechanisms by which Apolipoprotein E (APOE) contributes to Alzheimer’s disease (AD) risk. Neurobiol. Dis. 138, 104788 (2020).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Li, K., Wang, G., Andersen, T., Zhou, P. & Pu, W. T. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PLoS ONE 9, e105779 (2014).
Ren, C. et al. Dual-reporter surrogate systems for efficient enrichment of genetically modified cells. Cell. Mol. Life Sci. 72, 2763–2772 (2015).
Standage-Beier, K. et al. A transient reporter for editing enrichment (TREE) in human cells. Nucleic Acids Res. 47, e120 (2019).
Brookhouser, N. et al. A Cas9-mediated adenosine transient reporter enables enrichment of ABE-targeted cells. BMC Biol. 18, 193 (2020).
Kim, J., Koo, B.-K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
Kobayashi, Y. et al. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 9, 13 (2017).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018).
van Rheenen, W., Peyrot, W. J., Schork, A. J., Lee, S. H. & Wray, N. R. Genetic correlations of polygenic disease traits: from theory to practice. Nat. Rev. Genet. 20, 567–581 (2019).
Carlson-Stevermer, J. & Saha, K. Genome editing in human pluripotent stem cells. Methods Mol. Biol. 1590, 165–174 (2017).
Cheng, T.-L. et al. Expanding C-T base editing toolkit with diversified cytidine deaminases. Nat. Commun. 10, 3612 (2019).
Jeong, Y. K., Song, B. & Bae, S. Current status and challenges of DNA base editing tools. Mol. Ther. 28, 1938–1952 (2020).
Zhang, W. et al. A high-throughput small molecule screen identifies farrerol as a potentiator of CRISPR/Cas9-mediated genome editing. eLife 9, (2020).
Li, W. et al. Rational design of small molecules to enhance genome editing efficiency by selectively targeting distinct functional states of CRISPR-Cas12a. Bioconjug. Chem. 31, 542–546 (2020).
Rees, H. A., Yeh, W.-H. & Liu, D. R. Development of hRad51-Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat. Commun. 10, 2212 (2019).
Coelho, M. A. et al. BE-FLARE: a fluorescent reporter of base editing activity reveals editing characteristics of APOBEC3A and APOBEC3B. BMC Biol. 16, 150 (2018).
Katti, A. et al. GO: a functional reporter system to identify and enrich base editing activity. Nucleic Acids Res. 48, 2841–2852 (2020).
St Martin, A. et al. A fluorescent reporter for quantification and enrichment of DNA editing by APOBEC-Cas9 or cleavage by Cas9 in living cells. Nucleic Acids Res. 46, e84 (2018).
Martin, A. S. et al. A panel of eGFP reporters for single base editing by APOBEC-Cas9 editosome complexes. Sci. Rep. 9, 497 (2019).
Huang, T. P. et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 37, 626–631 (2019).
Jeong, Y. K., Yu, J. & Bae, S. Construction of non-canonical PAM-targeting adenosine base editors by restriction enzyme-free DNA cloning using CRISPR-Cas9. Sci. Rep. 9, 4939 (2019).
Kang, S.-H. et al. Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment. Nat. Commun. 11, 3596 (2020).
Stemmer, M., Thumberger, T., Del Sol Keyer, M., Wittbrodt, J. & Mateo, J. L. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 10, e0124633 (2015).
Manghwar, H. et al. CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off-target evaluation, and strategies to mitigate off-target effects. Adv. Sci. 7, 1902312 (2020).
Labuhn, M. et al. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res. 46, 1375–1385 (2018).
Park, S. & Beal, P. A. Off-target editing by CRISPR-guided DNA base editors. Biochemistry 58, 3727–3734 (2019).
Lee, H. K. et al. Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat. Commun. 9, 4804 (2018).
Zischewski, J., Fischer, R. & Bortesi, L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol. Adv. 35, 95–104 (2017).
Crosetto, N. et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365 (2013).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Wienert, B., Wyman, S. K., Yeh, C. D., Conklin, B. R. & Corn, J. E. CRISPR off-target detection with DISCOVER-seq. Nat. Protoc. 15, 1775–1799 (2020).
Liang, P. et al. Effective gene editing by high-fidelity base editor 2 in mouse zygotes. Protein Cell 8, 601–611 (2017).
Xu, W. et al. Multiplex nucleotide editing by high-fidelity Cas9 variants with improved efficiency in rice. BMC Plant Biol. 19, 511 (2019).
Liu, G., Zhang, Y. & Zhang, T. Computational approaches for effective CRISPR guide RNA design and evaluation. Comput. Struct. Biotechnol. J. 18, 35–44 (2020).
Huang, T. P., Newby, G. A. & Liu, D. R. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat. Protoc. https://doi.org/10.1038/s41596-020-00450-9 (2021).
Chang, Y.-J. et al. CRISPR base editing in induced pluripotent stem Cells. Methods Mol. Biol. 2045, 337–346 (2019).
Lino, C. A., Harper, J. C., Carney, J. P. & Timlin, J. A. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 25, 1234–1257 (2018).
Ortmann, D. & Vallier, L. Variability of human pluripotent stem cell lines. Curr. Opin. Genet. Dev. 46, 179–185 (2017).
McGrath, E. et al. Targeting specificity of APOBEC-based cytosine base editor in human iPSCs determined by whole genome sequencing. Nat. Commun. 10, 5353 (2019).
Baghbaderani, B. A. et al. Detailed characterization of human induced pluripotent stem cells manufactured for therapeutic applications. Stem Cell Rev. Rep. 12, 394–420 (2016).
Wang, X. et al. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotechnol. 36, 946–949 (2018).
Acknowledgements
Funding for this work was provided by the National Institutes of Health (R01GM121698 to D.A.B, R21AG056706 to D.A.B, R01GM106081 to X.W.) and the Arizona Biomedical Research Commission (ADHS16-162401 to D.A.B). N.B. was supported by a fellowship from the International Foundation for Ethical Research. We would like to thank the ASU Biodesign Flow Cytometry core for assistance with flow-cytometry-related experiments.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to this work. S.J.T, K.S.B., N.B. and D.A.B. wrote the paper. K.S.B. generated figures. S.J.T. and N.B. designed sample data. X.W. and D.A.B. oversaw the writing and layout of the manuscript. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.J.T., N.B., K.S.B., X.W. and D.A.B. are coinventors on a provisional patent application.
Additional information
Peer review information Nature Protocols thanks Krishanu Saha and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related Links
Key references using this protocol
Standage-Beier, K. et al. Nucl. Acids Res. 47, e120 (2019): https://doi.org/10.1093/nar/gkz713
Brookhouser, N. et al. Stem Cell Reports. 14, 184–191 (2020): https://doi.org/10.1016/j.stemcr.2019.12.013
Brookhouser, N. et al. BMC Biol. 18, 193 (2020): https://doi.org/10.1186/s12915-020-00929-7
Supplementary information
Rights and permissions
About this article
Cite this article
Tekel, S.J., Brookhouser, N., Standage-Beier, K. et al. Cytosine and adenosine base editing in human pluripotent stem cells using transient reporters for editing enrichment. Nat Protoc 16, 3596–3624 (2021). https://doi.org/10.1038/s41596-021-00552-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00552-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.