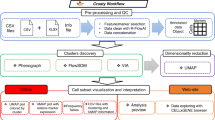
Abstract
The dimensionality of cytometry data has strongly increased in the last decade, and in many situations the traditional manual downstream analysis becomes insufficient. The field is therefore slowly moving toward more automated approaches, and in this paper we describe the protocol for analyzing high-dimensional cytometry data using FlowSOM, a clustering and visualization algorithm based on a self-organizing map. FlowSOM is used to distinguish cell populations from cytometry data in an unsupervised way and can help to gain deeper insights in fields such as immunology and oncology. Since the original FlowSOM publication (2015), we have validated the tool on a wide variety of datasets, and to write this protocol, we made use of this experience to improve the user-friendliness of the package (e.g., comprehensive functions replacing commonly required scripts). Where the original paper focused mainly on the algorithm description, this protocol offers user guidelines on how to implement the procedure, detailed parameter descriptions and troubleshooting recommendations. The protocol provides clearly annotated R code, and is therefore relevant for all scientists interested in computational high-dimensional analyses without requiring a strong bioinformatics background. We demonstrate the complete workflow, starting from data preparation (such as compensation, transformation and quality control), including detailed discussion of the different FlowSOM parameters and visualization options, and concluding with how the results can be further used to answer biological questions, such as statistical comparison between groups of interest. An average FlowSOM analysis takes 1–3 h to complete, though quality issues can increase this time considerably.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The flow cytometry data files and additional files are publicly available on FlowRepository (https://flowrepository.org/id/FR-FCM-ZZQY) and (https://flowrepository.org/id/FR-FCM-Z2TQ).
Code availability
The FlowSOM code is publicly available on GitHub (https://github.com/saeyslab/FlowSOM) and on Bioconductor (https://bioconductor.org/packages/release/bioc/html/FlowSOM.html). A demo script including the protocol that was described here and the source code for the figures in this paper are available on GitHub (https://github.com/saeyslab/FlowSOM_protocol). A list of the used R libraries and the versions can be found in Supplementary Note 1.
References
Adan, A., Alizada, G., Kiraz, Y., Baran, Y. & Nalbant, A. Flow cytometry: basic principles and applications. Crit. Rev. Biotechnol. 37, 163–176 (2017).
Liechti, T. & Roederer, M. OMIP-051 – 28-color flow cytometry panel to characterize B cells and myeloid cells. Cytometry A 95, 150–155 (2019).
Spitzer, M. H. & Nolan, G. P. Mass cytometry: single cells, many features. Cell 165, 780–791 (2016).
Futamura, K. et al. Novel full-spectral flow cytometry with multiple spectrally-adjacent fluorescent proteins and fluorochromes and visualization of in vivo cellular movement. Cytometry A 87, 830–842 (2015).
Saeys, Y., Van Gassen, S. & Lambrecht, B. N. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 16, 449–462 (2016).
Van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015).
Emmaneel, A. et al. A computational pipeline for the diagnosis of CVID patients. Front. Immunol. 10, 2009 (2019).
Guilliams, M. et al. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity 45, 669–684 (2016).
Ellis, B. et al. flowCore: flowCore: basic structures for flow cytometry data. Bioconductor version: release (3.11). https://doi.org/10.18129/B9.bioc.flowCore (2020).
Kohonen, T. The self-organizing map. Proc. IEEE 78, 1464–1480 (1990).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Shekhar, K., Brodin, P., Davis, M. M. & Chakraborty, A. K. Automatic classification of cellular expression by nonlinear stochastic embedding (ACCENSE). Proc. Natl Acad. Sci. USA 111, 202–207 (2014).
Aghaeepour, N., Nikolic, R., Hoos, H. H. & Brinkman, R. R. Rapid cell population identification in flow cytometry data. Cytometry A 79A, 6–13 (2011).
Lo, K., Hahne, F., Brinkman, R. R. & Gottardo, R. flowClust: a Bioconductor package for automated gating of flow cytometry data. BMC Bioinformatics 10, 145 (2009).
Ye, X. & Ho, J. W. K. Ultrafast clustering of single-cell flow cytometry data using FlowGrid. BMC Syst. Biol. 13, 35 (2019).
Weber, L. M. & Robinson, M. D. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytometry A 89, 1084–1096 (2016).
Liu, X. et al. A comparison framework and guideline of clustering methods for mass cytometry data. Genome Biol. 20, 297 (2019).
Pedersen, C. B. & Olsen, L. R. Algorithmic clustering of single-cell cytometry data—how unsupervised are these analyses really? Cytometry A 97, 219–221 (2020).
Liu, P. et al. Recent advances in computer-assisted algorithms for cell subtype identification of cytometry data. Front. Cell Dev. Biol. 8, 234 (2020).
Hamers, A. A. J. et al. Human monocyte heterogeneity as revealed by high-dimensional mass cytometry. Arterioscler. Thromb. Vasc. Biol. 39, 25–36 (2019).
Kratochvíl, M., Bednárek, D., Sieger, T., Fišer, K. & Vondrášek, J. ShinySOM: graphical SOM-based analysis of single-cell cytometry data. Bioinformatics https://doi.org/10.1093/bioinformatics/btaa091 (2020).
Weber, L. M., Nowicka, M., Soneson, C. & Robinson, M. D. diffcyt: differential discovery in high-dimensional cytometry via high-resolution clustering. Commun. Biol. 2, 183 (2019).
Van Gassen, S., Gaudilliere, B., Angst, M. S., Saeys, Y. & Aghaeepour, N. CytoNorm: a normalization algorithm for cytometry data. Cytometry A 97, 268–278 (2020).
Kotecha, N., Krutzik, P. O. & Irish, J. M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 53, 10.17.1–10.17.24 (2010).
FlowJoTM Software (Becton, Dickinson, 2019).
Amir, E. D. et al. Development of a comprehensive antibody staining database using a standardized analytics pipeline. Front. Immunol. 10, 1315 (2019).
Lacombe, F., Lechevalier, N., Vial, J. P. & Béné, M. C. An R-derived FlowSOM process to analyze unsupervised clustering of normal and malignant human bone marrow classical flow cytometry data. Cytometry A 95, 1191–1197 (2019).
Bhattacharya, S. et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 5, 180015 (2018).
Vetters, J. et al. The ubiquitin-editing enzyme A20 controls NK cell homeostasis through regulation of mTOR activity and TNF. J. Exp. Med. 216, 2010–2023 (2019).
Spidlen, J., Breuer, K., Rosenberg, C., Kotecha, N. & Brinkman, R. R. FlowRepository: a resource of annotated flow cytometry datasets associated with peer-reviewed publications. Cytometry A 81A, 727–731 (2012).
Roca, C. P. et al. AutoSpill: a method for calculating spillover coefficients in high-parameter flow cytometry. Preprint at bioRxiv https://doi.org/10.1101/2020.06.29.177196 (2020).
Finak, G., Jiang, W. & Gottardo, R. CytoML for cross-platform cytometry data sharing. Cytometry A 93, 1189–1196 (2018).
Finak, G. & Jiang, M. flowWorkspace: infrastructure for representing and interacting with gated and ungated cytometry data sets (2020).
Emmaneel, A. PeacoQC: peak-based selection of high quality cytometry data. https://github.com/saeyslab/PeacoQC (2020).
Finak, G. et al. OpenCyto: an open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS Comput. Biol. 10, e1003806 (2014).
Malek, M. et al. flowDensity: reproducing manual gating of flow cytometry data by automated density-based cell population identification. Bioinformatics 31, 606–607 (2015).
Nowicka, M. et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Research 6, 748 (2019).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Pelon, F. et al. Cancer-associated fibroblast heterogeneity in axillary lymph nodes drives metastases in breast cancer through complementary mechanisms. Nat. Commun. 11, 404 (2020).
Duetz, C., Bachas, C., Westers, T. M. & van de Loosdrecht, A. A. Computational analysis of flow cytometry data in hematological malignancies: future clinical practice? Curr. Opin. Oncol. 32, 162–169 (2020).
Ho, W. J. et al. Multipanel mass cytometry reveals anti–PD-1 therapy–mediated B and T cell compartment remodeling in tumor-draining lymph nodes. JCI Insight 5, e132286 (2020).
Metelli, A. et al. Thrombin contributes to cancer immune evasion via proteolysis of platelet-bound GARP to activate LTGF-β. Sci. Transl. Med. 12, eaay4860 (2020).
Shaul, M. E. et al. Circulating neutrophil subsets in advanced lung cancer patients exhibit unique immune signature and relate to prognosis. FASEB J. 34, 4204–4218 (2020).
Laban, K. G. et al. cDC2 and plasmacytoid dendritic cells diminish from tissues of patients with non-Hodgkin orbital lymphoma and idiopathic orbital inflammation. Eur. J. Immunol. 50, 548–557 (2020).
Ho, W. J. et al. Viral status, immune microenvironment and immunological response to checkpoint inhibitors in hepatocellular carcinoma. J. Immunother. Cancer 8, e000394 (2020).
Yarchoan, M. et al. Effects of B cell–activating factor on tumor immunity. JCI Insight 5, e136417 (2020).
Ghorani, E. et al. The T cell differentiation landscape is shaped by tumour mutations in lung cancer. Nat. Cancer 1, 546–561 (2020).
Friebel, E. et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell 181, 1626–1642.e20 (2020).
Perez, C. et al. Immunogenomic identification and characterization of granulocytic myeloid-derived suppressor cells in multiple myeloma. Blood 136, 199–209 (2020).
Kverneland, A. H. et al. Adoptive cell therapy in combination with checkpoint inhibitors in ovarian cancer. Oncotarget 11, 2092–2105 (2020).
Färkkilä, A. et al. Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat. Commun. 11, 1459 (2020).
Ji, A. L. et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 497–514.e22 (2020).
Khalsa, J. K. et al. Immune phenotyping of diverse syngeneic murine brain tumors identifies immunologically distinct types. Nat. Commun. 11, 3912 (2020).
Ali, H. R. et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Cancer 1, 163–175 (2020).
Ranganath, T. et al. Characterization of the impact of daclizumab beta on circulating natural killer cells by mass cytometry. Front. Immunol. 11, 714 (2020).
Grayson, J. M. et al. Photodepletion with 2-Se-Cl prevents lethal graft-versus-host disease while preserving antitumor immunity. PLoS ONE 15, e0234778 (2020).
Leelatian, N. et al. Unsupervised machine learning reveals risk stratifying glioblastoma tumor cells. eLife 9, e56879 (2020).
Gudbergsson, J. M. et al. Conventional treatment of glioblastoma reveals persistent CD44+ subpopulations. Mol. Neurobiol. 57, 3943–3955 (2020).
Wuggenig, P. et al. Loss of the branched-chain amino acid transporter CD98hc alters the development of colonic macrophages in mice. Commun. Biol. 3, 130 (2020).
Utz, S. G. et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell 181, 557–573.e18 (2020).
Hawke, L. G., Mitchell, B. Z. & Ormiston, M. L. TGF-β and IL-15 synergize through MAPK pathways to drive the conversion of human NK cells to an innate lymphoid cell 1–like phenotype. J. Immunol. 204, 3171–3181 (2020).
Hagert, C. F., Bohn, A. B., Wittenborn, T. R. & Degn, S. E. Seeing the confetti colors in a new light utilizing flow cytometry and imaging flow cytometry. Cytometry A 97, 811–823 (2020).
Rein, I. D., Notø, H. Ø., Bostad, M., Huse, K. & Stokke, T. Cell cycle analysis and relevance for single-cell gating in mass cytometry. Cytometry A 97, 832–844 (2020).
Jokela, H. et al. Fetal-derived macrophages persist and sequentially maturate in ovaries after birth in mice. Eur. J. Immunol. 50, 1500–1514 (2020).
Grandi, F. C. et al. Single-cell mass cytometry reveals cross-talk between inflammation-dampening and inflammation-amplifying cells in osteoarthritic cartilage. Sci. Adv. 6, eaay5352 (2020).
Neeland, M. R. et al. Mass cytometry reveals cellular fingerprint associated with IgE+ peanut tolerance and allergy in early life. Nat. Commun. 11, 1091 (2020).
Jang, J. S. et al. Single-cell mass cytometry on peripheral blood identifies immune cell subsets associated with primary biliary cholangitis. Sci. Rep. 10, 12584 (2020).
Eichmann, M. et al. Costimulation blockade disrupts CD4+ T cell memory pathways and uncouples their link to decline in β-cell function in type 1 diabetes. J. Immunol. 204, 3129–3138 (2020).
Muppidi, A. & Radfar, M. Löfgren’s syndrome sarcoidosis and Non-LS sarcoidosis prediction using 1d-Convolutional neural networks. Inform. Med. Unlocked 19, 100328 (2020).
Mitsialis, V. et al. Single-cell analyses of colon and blood reveal distinct immune cell signatures of ulcerative colitis and Crohn’s disease. Gastroenterology https://doi.org/10.1053/j.gastro.2020.04.074 (2020).
Brooks, A. E. S. et al. Ex vivo human adipose tissue derived mesenchymal stromal cells (ASC) are a heterogeneous population that demonstrate rapid culture-induced changes. Front. Pharmacol. 10, 1695 (2020).
Johnson, B. Z. et al. Pediatric burn survivors have long-term immune dysfunction with diminished vaccine response. Front. Immunol. 11, 1481 (2020).
Eccles, J. D. et al. T-bet+ memory B cells link to local cross-reactive IgG upon human rhinovirus infection. Cell Rep 30, 351–366.e7 (2020).
De Biasi, S. et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 11, 3434 (2020).
Zhao, N. Q. et al. Treated HIV infection alters phenotype but not HIV-specific function of peripheral blood natural killer cells. Front. Immunol. 11, 829 (2020).
Ma, T. et al. HIV efficiently infects T cells from the endometrium and remodels them to promote systemic viral spread. eLife 9, e55487 (2020).
Goshu, B. A., Chen, H., Moussa, M., Cheng, J. & Catalfamo, M. Combination rhIL-15 and anti-PD-L1 (Avelumab) enhances HIVGag-specific CD8 T-cell function. J. Infect. Dis. https://doi.org/10.1093/infdis/jiaa269 (2020).
Mathew, D. et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science https://doi.org/10.1126/science.abc8511 (2020).
Mersmann, O., Beleites, C., Hurling, R., Friedman, A. & Ulrich, J. M. microbenchmark: Accurate Timing Functions. https://rdrr.io/cran/microbenchmark/
Monaco, G. et al. flowAI: automatic and interactive anomaly discerning tools for flow cytometry data. Bioinformatics 32, 2473–2480 (2016).
Fletez‐Brant, K., Špidlen, J., Brinkman, R. R., Roederer, M. & Chattopadhyay, P. K. flowClean: automated identification and removal of fluorescence anomalies in flow cytometry data. Cytometry A 89, 461–471 (2016).
Finak, G. et al. High-throughput flow cytometry data normalization for clinical trials. Cytometry A 85A, 277–286 (2014).
Hahne, F. et al. Per-channel basis normalization methods for flow cytometry data. Cytometry A 77A, 121–131 (2009).
Rybakowska, P., Alarcón-Riquelme, M. E. & Marañón, C. Key steps and methods in the experimental design and data analysis of highly multi-parametric flow and mass cytometry. Comput. Struct. Biotechnol. J. 18, 874–886 (2020).
Stanley, N. et al. VoPo leverages cellular heterogeneity for predictive modeling of single-cell data. Nat. Commun. 11, 3738 (2020).
Acknowledgements
S.V.G. is an ISAC Marylou Ingram Scholar and supported by an FWO postdoctoral research grant (Research Foundation—Flanders). We thank the VIB Flow Core for training, support and access to the instrument park. This research received funding from the Flemish Government (AI Research program). A.E. is supported by the PID Grand Challenges Program of VIB. This VIB Program received support from the Flemish Government under the Management Agreement 2017–2021 (VR 2016 2312 Doc.1521/4).
Author information
Authors and Affiliations
Contributions
S.V.G and Y.S. conceptualized the FlowSOM algorithm. S.V.G., A.C., A.E. and K.Q. wrote the FlowSOM code. S.V.G., Y.S. and J.A. supervised the work. K.Q. wrote the manuscript. All authors edited, read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Nima Aghaeepour, Étienne Becht and Enrico Lugli for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Van Gassen, S. et al. Cytometry A 87, 636–645 (2015): https://doi.org/10.1002/cyto.a.22625
Guilliams, M. et al. Immunity 45, 669–684 (2016): https://doi.org/10.1016/j.immuni.2016.08.015
Emmaneel, A. et al. Front. Immunol. 10, 2009 (2019): https://doi.org/10.3389/fimmu.2019.02009
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Supplementary Note.
Rights and permissions
About this article
Cite this article
Quintelier, K., Couckuyt, A., Emmaneel, A. et al. Analyzing high-dimensional cytometry data using FlowSOM. Nat Protoc 16, 3775–3801 (2021). https://doi.org/10.1038/s41596-021-00550-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00550-0
This article is cited by
-
CytoPipeline and CytoPipelineGUI: a Bioconductor R package suite for building and visualizing automated pre-processing pipelines for flow cytometry data
BMC Bioinformatics (2024)
-
SpiDe-Sr: blind super-resolution network for precise cell segmentation and clustering in spatial proteomics imaging
Nature Communications (2024)
-
Bifidobacterium infantis supplementation versus placebo in early life to improve immunity in infants exposed to HIV: a protocol for a randomized trial
BMC Complementary Medicine and Therapies (2023)
-
An end-to-end workflow for multiplexed image processing and analysis
Nature Protocols (2023)
-
The extrafollicular B cell response is a hallmark of childhood idiopathic nephrotic syndrome
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.