Abstract
The coming of the Last Universal Cellular Ancestor (LUCA) was the singular watershed event in the making of the biotic world. If the coming of LUCA marked the crossing of the “Darwinian Threshold”, then pre-LUCA evolution must have been Pre-Darwinian and at least partly non-Darwinian. But how did Pre-Darwinian evolution before LUCA actually operate? I broaden our understanding of the central mechanism of biological evolution (i.e., variation-selection-inheritance) and then extend this broadened understanding to its natural starting point: the origin(s) of the First Universal Cellular Ancestors (FUCAs) before LUCA. My hypothesis centers upon vesicles’ making-and-remaking as variation and competition as selection. More specifically, I argue that vesicles’ acquisition and merger, via breaking-and-repacking, proto-endocytosis, proto-endosymbiosis, and other similar processes had been a central force of both variation and selection in the pre-Darwinian epoch. These new perspectives shed important new light upon the origin of FUCAs and their subsequent evolution into LUCA.


Similar content being viewed by others
Notes
E.g., Oparin 1953 [1938]; Fox 1973; Woese and Fox 1977; Gánti 1997; Szathmáry and Maynard Smith 1997; Norris and Raine 1998; Woese 1998, 2000, 2002, 2004; Cavalier-Smith 2001; Szostak et al. 2001; Luisi 2006, 2016; Chen and Walde 2010; Fry 2011; Pross 2011; Egel 2012, 2017; Bouchard 2014; Doolittle 2014; Koonin 2014a, b; O’Malley 2014; Baum 2015; Damer and Deamer 2015, 2020; Spitzer et al. 2015; Smith and Morowitz 2016; Pascal and Pross 2016; Cornish-Bowden and Cárdenas 2017; Garson 2017; Lanier and Williams 2017; Spitzer 2017; Toman and Flegr 2017; Doolittle and Inkpen 2018; Deamer 2019; Deamer et al. 2019; Lopez and Fiore 2019; Kunnev 2020. I apologize for not citing all the relevant literature: it is simply too voluminous. For overviews of our journey since Oparin, see Miller et al. 1997; Lazcano 2010; Deamer 2019.
The notation of FUCA is similar to the First Eukaryotic Common Ancestors (FECAs) before the Last Eukaryotic Common Ancestor (LECA). Just as FECAs were most likely a pool of organisms (Eme et al. 2017), FUCAs were also most likely a pool of (proto-)organisms. See Section I below.
I also skip some rather minor quibbles (e.g., whether LUCA had introns).
Hence, my position critically differs from Prosdocimi et al. (2019), who also used the abbreviation FUCA for “the First Universal Common Ancestor”. Prosdocimi et al. insists that FUCA is not a cell but a RNA-dependent peptide-synthesis machinery or peptidyl transferase center. Also, they merely hypothesized that FUCA existed but provided no hypothesis for its origin.
Indeed, Vetsigian et al. (2006, p. 10,697) came extremely close to such a position: “The central conjecture in our model is that innovation-sharing, which involves horizontal transfer of genes and perhaps other complex elements among the evolving entities [a dynamic far more rampant and pervasive than our current perception of horizontal gene transfer (HGT)], is required to bring the evolving translation apparatus, its code, and by implication the cell itself to their current condition.” (Italics added.) Moreover, starting with a quite different perspective, Woese and Fox (1977, p. 5) had actually argued that “endosymbiosis should probably be considered an aboriginal (…) [and not-so-rare] trait” rather than “an acquired trait” or “a relatively recent and rare occurrence”. For this author, it is a mystery why Woese’s penetrating mind had not joined these two themes together.
Alternatively, if there had been more than two sub-populations initially, eventually only two had survived.
This possibility best explains the great diversity of this group of virus, and why ( +)ssRNA viruses remain the largest group of viruses (de Farias et al. 2017).
By biological-functional, we mean that the same biological function can be performed by different molecules, with or without significant structural or sequence similarities. For example, an alpha-helix can be formed by different stretches of peptide.
The evolution of the standard (or universal) genetic code is a research area all by itself. Here, suffice to say that our thesis is compatible with the emerging consensus that three mechanisms must have jointly played key role in the evolution of the code and they reinforce each other (see Koonin and Novozhilov 2017 for review). The three key mechanisms are: stereochemistry, coevolution of the code and then translation plus proteins, and error minimization. Note, however, all three mechanisms implicitly center on the evolution of the code within a single cell population and this possibility is highly unlikely (see below). Hence, HBMT as a mechanism that can draw from “global innovation” [and subsumes HGT (e.g., Vetsigian et al. 2006)] may be absolutely necessary.
The fact that replicons with protein capsids (i.e., viruses) are more widespread than replicons without protein capsids (i.e., viroids) also suggest that the former is more advantageous than the latter. Also, viroids have so far only been found in higher plants whereas viruses have been found in all three domains. This fact suggests that viroids most likely have a recent origin.
This possibility therefore supports the “TAGTF” thesis while going against the “hydrothermal alkaline vents (HTAV)” thesis. See below.
Megaviruses, as double-stranded DNA viruses, must have originated after the origin of DNA replication. Hence, megaviruses do not support the thesis that early replicons or protocells had possessed big genomes.
So far, no Hsp12 homologs have been discovered beyond fungi, but this may be due to the fact that membrane proteins are less conserved than water-solvable proteins (Sojo et al. 2016). More speculatively, polyglycine, made of Glycine, might have played a similar role in stabilizing protocell membranes (Kim et al. 2019).
To resolve this classic chicken-and-egg difficulty, Koonin wisely switched from the thesis of maritime origin within inorganic chambers (Koonin and Martin 2005; Koonin 2009) to a thesis of terrestrial origin (Mulkidjanian et al. 2009, 2012; see also Koonin 2014a, b). Yet, Koonin (2014b) has retained the possibility of multiple escape from the communal ancestors (see also Koonin and Dolja 2013). See below.
Hence, by 1998, Woese had changed from his original conception of progenotes that did not differentiate progenotes and genotes to a more valid conception that does differentiate the two.
Indeed, multiple escapes and multiple invasions are like two sides of the same miracle.
Our testing of this possibility with computer simulation will be reported separately.
Indeed, UBLs play critical roles in pre-mRNA splicing in eukaryotes (Chanarat and Mishra 2018).
Abbreviations
- HBMT:
-
Horizontal biomolecules transfer
- HGT:
-
Horizontal gene transfer
- HTAV:
-
Hydrothermal alkaline vent
- TAGTF:
-
Terrestrial anoxic geothermal fields
References
Adamala K, Szostak JW (2013) Competition between model protocells driven by an encapsulated catalyst. Nat Chem 5:495–501
Altan-Bonnet N (2017) Lipid tales of viral replication and transmission. Trends Cell Biol 27:201–213
Armstrong DL, Lancet D, Zidovetzki R (2018) Replication of simulated prebiotic amphiphilic vesicles in a finite environment exhibits complex behavior that includes high progeny variability and competition. Astrobiology 18:419–430
Baum DA (2015) Selection and the origin of cells. Bioscience 65:678–684
Bedau MA, Cleland CE (2010) The nature of life: classical and contemporary perspectives from philosophy and science. Cambridge University Press, Cambridge
Black RA, Blosser MC, Stottrup BL, Tavakley R, Deamer DW, Keller SL (2013) Nucleobases bind to and stabilize aggregates of a prebiotic amphiphile, providing a viable mechanism for the emergence of protocells. Proc Natl Acad Sci USA 110:13272–13276
Booth A, Doolittle WF (2015) Eukaryogenesis, how special really? Proc Natl Acad Sci USA 112:10278–10285
Bouchard F (2014) Ecosystem evolution is about variation and persistence, not populations and reproduction. Biol Theory 9:382–391
Brack A (1998) The molecular origins of life: assembling pieces of the puzzle. Cambridge University Press, Cambridge
Budin I, Szostak JW (2011) Physical effects underlying the transition from primitive to modern cell membranes. Proc Natl Acad Sci USA 108:5249–5254
Budin I, Debnath A, Szostak JW (2012) Concentration-driven growth of model protocell membranes. J Am Chem Soc 134:20812–20819
Burroughs AM, Iyer LM, Aravind L (2012a) The natural history of ubiquitin and ubiquitin-related domains. Front Biosci 17:1433–1460
Burroughs AM, Iyer LM, Aravind L (2012b) Structure and evolution of ubiquitin and ubiquitin-related domains. Methods Mol Biol 832:15–63
Caforio A, Siliakus MF, Exterkate M, Jain S, Jumde VR, Andringa RLH, Kengen SWM, Minnaard AJ, Driessen AJM, van der Oost J (2018) Converting Escherichia coli into an archaebacterium with a hybrid heterochiral membrane. Proc Natl Acad Sci USA 115:3704–3709
Cantine MD, Fournier GP (2018) Environmental adaptation from the origin of life to the last universal common ancestor. Orig Life Evol Biosph 48:35–54
Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, Brar GA, Weissman JS, Regev A, Thierry-Mieg N, Cusick ME, Vidal M (2012) Proto-genes and de novo gene birth. Nature 487:370–374
Cavalier-Smith T (2001) Obcells as proto-organisms: membrane heredity, lithophosphorylation, and the origins of the genetic code, the first cells, and photosynthesis. J Mol Evol 53:555–595
Chanarat S, Mishra SK (2018) Emerging roles of ubiquitin-like proteins in pre-mRNA splicing. Trends Biochem Sci 43:896–907
Charlebois RL, Doolittle WF (2004) Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res 14:2469–2477
Chen IA, Walde P (2010) From self-assembled vesicles to protocells. Cold Spring Harb Perspect Biol 2:a002170
Chen IA, Roberts RW, Szostak JW (2004) The emergence of competition between model protocells. Science 305:1474–1476
Cheng Z, Luisi PL (2003) Coexistence and mutual competition of vesicles with different size distributions. J Phys Chem B 107:10940–10945
Coleman GA, Pancost RD, Williams TA (2019) Investigating the origins of membrane phospholipid biosynthesis genes using outgroup-free rooting. Genome Biol Evol 11:883–898
Cornell CE, Black RA, Xue M, Litz HE, Ramsay A, Gordon M, Mileant A, Cohen ZR, Williams JA, Lee KK, Drobny GP, Keller SL (2019) Prebiotic amino acids bind to and stabilize prebiotic fatty acid membranes. Proc Natl Acad Sci USA 116:17239–17244
Cornish-Bowden A, Cárdenas ML (2017) Life before LUCA. J Theor Biol 434:68–74
Da Silva L, Maurel MC, Deamer D (2015) Salt-promoted synthesis of RNA-like molecules in simulated hydrothermal conditions. J Mol Evol 80:86–97
Dacks JB, Field MC, Buick R, Eme L, Gribaldo S, Roger AJ, Brochier-Armanet C, Devos DP (2016) The changing view of eukaryogenesis-fossils, cells, lineages and how they all come together. J Cell Sci 129:3695–3703
Damer B, Deamer D (2015) Coupled phases and combinatorial selection in fluctuating hydrothermal pools: a scenario to guide experimental approaches to the origin of cellular life. Life 5:872–887
Damer B, Deamer D (2020) The hot spring hypothesis for an origin of life. Astrobiology 20:429–452
de Duve C (2005a) The onset of natural selection. Nature 433:581–582
de Duve C (2005b) Singularities: landmarks on the pathways of life. Cambridge University Press, Cambridge
de Farias ST, dos Santos JAP, Regro TG, Jose MV (2017) Origin and evolution of RNA-dependent RNA polymerase. Front Genet. https://doi.org/10.3389/fgene.2017.00125
Deamer DW (2019) Assembling life. Oxford University Press, Oxford
Deamer DW, Barchfeld GL (1982) Encapsulation of macromolecules by lipid vesicles under simulated prebiotic conditions. J Mol Evol 18:203–206
Deamer D, Damer B, Kompanichenko V (2019) Hydrothermal chemistry and the origin of cellular life. Astrobiology 19:1523–1537
Doolittle WF (2000) The nature of the universal ancestor and the evolution of the proteome. Curr Opin Struct Biol 10:355–358
Doolittle WF (2014) Natural selection through survival alone, and the possibility of Gaia. Biol Philos 29:415–423
Doolittle WF, Inkpen SA (2018) Processes and patterns of interaction as units of selection: an introduction to ITSNTS thinking. Proc Natl Acad Sci USA 115:4006–4014
Egel R (2009) Peptide-dominated membranes preceding the genetic takeover by RNA: latest thinking on a classic controversy. BioEssays 31:1100–1109
Egel R (2012) Life’s order, complexity, organization, and its thermodynamic-holistic imperatives. Life 2:323–363
Egel R (2017) ‘Parabiotic evolution’: from stochasticity in geochemical and subsequent processes to genes, genomes and modular cells. Preprints 2017100153. https://doi.org/10.20944/preprints201710.0153.v1
El Baidouri F, Venditti C, Suzuki S, Meade A, Humpries S (2020) Phenotypic reconstruction of the last universal common ancestor reveals a complex cell. bioRxiv. https://doi.org/10.1101/2020.08.20.260398
Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG (2017) Archaea and the origin of eukaryotes. Nat Rev Microbiol 15:711–723
Forterre P (2006) Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc Natl Acad Sci USA 103:3669–3674
Forterre P, Philippe H (1999) Where is the root of the universal tree of life? BioEssays 21:871–879
Fournier GP, Andam CP, Gogarten JP (2015) Ancient horizontal gene transfer and the last common ancestors. BMC Evol Biol 15:70
Fox SW (1973) Molecular evolution to the first cells. Pure Appl Chem 34:641
Fox GE (2010) Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol 2:a003483. https://doi.org/10.1101/cshperspect.a003483
Fox GE, Luehrsen KR, Woese CR (1982) Archaebacterial 5S Ribosomal RNA. Zbl Bakt Hyg. i. Abr Orig C 3:330–345
Fox GE, Tran Q, Yonath A (2012) An exit cavity was crucial to the polymerase activity of the early ribosome. Astrobiology 12:57–60
Francis BR (2011) An alternative to the RNA world hypothesis. Trends Evol Biol 3:e2
Francis BR (2013) Evolution of the genetic code by incorporation of amino acids that improved or changed protein function. J Mol Evol 77:134–158
Francis BR (2015) The hypothesis that the genetic code originated in coupled synthesis of proteins and the evolutionary predecessors of nucleic acids in primitive cells. Life 5:467–505
Frenkel-Pinter M, Haynes JW, Mohyeldin AM, Martin C, Sargon AB, Petrov AS, Krishnamurthy R, Hud NV, Williams LD, Leman LJ (2020) Mutually stabilizing interactions between proto-peptides and RNA. Nat Commun 11:3137
Fry I (2011) The role of natural selection in the origin of life. Orig Life Evol Biosph 41:3–16
Gánti T (1997) Biogenesis itself. J Theor Biol 187:583–593
Garson J (2017) A generalized selected effects theory of function. Phil Sci 84:523–543
Glansdorff N (2002) About the last common ancestor, the universal life-tree, and lateral gene transfer: a reappraisal. Mol Microbiol 38:177–185
Glansdorff N, Xu Y, Labedan B (2008) The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol Direct 3:29
Gogarten JP, Deamer D (2016) Is LUCA a thermophilic progenote? Nat Microbiol 1:16229
Goldman AD, Kacar B (2021) Cofactros are remnants of life’s origin and early evolution. J Mol Evol 89:127–133
Harold FM (2014) In search of cell history: the evolution of life’s building blocks. University of Chicago Press, Chicago
Harris AJ, Goldman AD (2021) The very early evolution of protein translocation across membranes. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1008623
Harris JK, Kelley ST, Spiegelman GB, Pace NR (2003) The genetic core of the universal ancestor. Genome Res 13:407–412
Higgs PG (2016) The effect of limited diffusion and wet-dry cycling on reversible polymerization reactions: implications for prebiotic synthesis of nucleic acids. Life 6:24
Higgs PG, Lehman N (2015) The RNA world: molecular cooperation at the origins of life. Nat Rev Genet 16:7–17
Hochstrasser M (2009) Origin and function of ubiquitin-like proteins. Nature 458:422–429
Horning DP, Joyce GF (2017) Amplification of RNA by an RNA polymerase ribozyme. Proc Natl Acad Sci USA 113:9786–9791
Ichihashi N, Yomo T (2014) Positive roles of compartmentalization in internal reactions. Curr Opin Chem Biol 22:12–17
Ichihashi N, Usui K, Kazuta Y, Sunami T, Matsuura T, Yomo T (2013) Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment. Nat Commun 4:2494. https://doi.org/10.1038/ncomms3494
Jackson JB (2016) Natural pH gradients in hydrothermal alkali vents were unlikely to have played a role in the origin of life. J Mol Evol 83:1–11
Jackson JB (2017) Ancient living organisms escaping from, or imprisoned in, the vents? Life 7:36
Jain R, Rivera MC, Lake JA (1999) Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA 96:3801–3806
Jain S, Caforio A, Driessen AJM (2014) Biosynthesis of archaeal membrane ether lipids. Front Microbiol 5:641
Joyce GF (2002) The antiquity of RNA-based evolution. Nature 418:214–221
Joyce GF, Szostak JW (2018) Protocells and RNA selfreplication. Cold Spring Harb Perspect Biol 10:a034801
Kandler O (1994a) The early diversification of life. In: Bengtson S (ed) Early life on earth: Nobel Symposium no. 84. Columbia University Press, New York, pp 152–160
Kandler O (1994b) Cell wall biochemistry in Archaea and its phylogenetic implications. J Biol Phys 20:165–169
Kandler O (1998) The early diversification of life and the origin of the three domains: a proposal. In: Wiegel J, Adams MWW (eds) Thermophiles: the keys to molecular evolution and the origin of life. Taylor & Francis, London, pp 19–28
Kim Y, Opron K, Burton ZF (2019) A tRNA- and anticodon-centric view of the evolution of aminoacyl-tRNA synthetases, tRNAomes, and the genetic code. Life 9:37. https://doi.org/10.3390/life9020037
Klein A, Bock M, Alt W (2017) Simple mechanisms of early life-simulation model on the origin of semi-cells. Biosystems 151:34–42
Koga Y (2011) Early evolution of membrane lipids: how did the lipid divide occur? J Mol Evol 72:274–282
Koga Y (2012) Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea 2012:789652
Koga Y (2014) From promiscuity to the lipid divide: on the evolution of distinct membranes in Archaea and Bacteria. J Mol Evol 78:234–242
Koonin EV (2003) Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol 1:127–136
Koonin EV (2009) On the origin of cells and viruses: primordial virus world scenario. Ann N Y Acad Sci 1178:47–64
Koonin EV (2014a) Carl Woese’s vision of cellular evolution and the domains of life. RNA Biol 11:197–204
Koonin EV (2014b) The origin of cellular life. Antonie Van Leeuwenhoek 106:27–41
Koonin EV (2016) Viruses and mobile elements as drivers of evolutionary transitions. Philos Trans R Soc Lond B Biol Sci 371:20150442
Koonin EV (2017) Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol Direct 12:5
Koonin EV, Martin W (2005) On the origin of genomes and cells within inorganic compartments. Trends Genet 21:647–654
Koonin EV, Dolja VV (2013) A virocentric perspective on the evolution of life. Curr Opin Virol 3:546–557
Koonin EV, Yutin N (2014) The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb Perspect Biol 6:a016188
Koonin EV, Makarova KS (2017) Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. Genome Biol Evol 9(10):2812–2825
Koonin EV, Novozhilov AS (2017) Origin and evolution of the universal genetic code. Annu Rev Genet 27:45–62
Koonin EV, Makarova KS, Wolf YI, Krupovic M (2020) Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat Rev Genet 21:119–131
Kovacs NA, Petrov AS, Lanier KA, Williams LD (2017) Frozen in time: the history of proteins. Mol Biol Evol 34:1252–1260
Krupovic M, Dolja VV, Koonin EV (2020) The LUCA and its complex virome. Nat Rev Microbiol 18:661–670
Ku C, Nelson-Sathi S, Roettger M, Sousa FL, Lockhart PJ, Bryant D, Hazkani-Covo E, McInerney JO, Landan G, Martin WF (2015) Endosymbiotic origins and differential loss of eukaryotic genes. Nature 254:427–432
Kunnev D (2020) Origin of life: the point of no return. Life 10:269. https://doi.org/10.3390/life10110269
Kurihara K, Okura Y, Matsuo M, Toyota T, Suzuki K, Sugawara T (2015) A recursive vesicle-based model protocell with a primitive model cell cycle. Nat Commun 6:8352
Lancet D, Shenhav B (2009) Compositional lipid protocells: reproduction without polynucleotides. In: Rasmussen S, Bedau MA, Chen L, Deamer D, Krakauer DC, Packard NH, Stadler PF (eds) Protocells: bridging nonliving and living matter. MIT, Cambridge, pp 233–252
Lane N, Martin WF (2015) Eukaryotes really are special, and mitochondria are why. Proc Natl Acad Sci USA 112:E4823
Lane N, Allen JF, Martin W (2010) How did LUCA make a living? Chemiosmosis in the origin of life. BioEssays 32:271–280
Lanier KA, Williams LD (2017) The origin of life: models and data. J Mol Evol 84:85–92
Lanier KA, Petrov AS, Williams LD (2017) The central symbiosis of molecular biology: molecules in mutualism. J Mol Evol 85:8–13
Lazcano A (2010) Historical development of origins research. Cold Spring Harb Perspect Biol 2:a002089
Lear JD, Wasserman ZR, DeGrado WF (1988) Synthetic amphiphilic peptide models for protein ion channels. Science 240:1177–1181
Leipe DD, Aravind L, Koonin EV (1999) Did DNA replication evolve twice independently? Nucleic Acids Res 27:3389–3401
Lombard J (2016) Early evolution of polyisoprenol biosynthesis and the origin of cell walls. PeerJ 4:e2626
Lombard J, López-García P, Moreira D (2012) The early evolution of lipid membranes and the three domains of life. Nat Rev Microbiol 10:507–515
Longo LM, Despotović D, Weil-Ktorza O, Walker MJ, Jabłońska J, Fridmann-Sirkis Y, Varani G, Metanis N, Tawfik DS (2020) Primordial emergence of a nucleic acid-binding protein via phase separation and statistical ornithine-to-arginine conversion. Proc Natl Acad Sci USA 117:15731–15739
López-García P, Moreira D (2015) Open questions on the origin of eukaryotes. Trends Ecol Evol 30:697–708
Lopez A, Fiore M (2019) Investigating prebiotic protocells for a comprehensive understanding of the origins of life: a prebiotic systems chemistry perspective. Life 9(2):49. https://doi.org/10.3390/life9020049
Luisi PL (2006) The emergence of life. Cambridge University Press, Cambridge
Luisi PL (2016) The emergence of life, 2nd edn. Cambridge University Press, Cambridge
Ma BG, Chen L, Ji HF, Chen ZH, Yang FR, Wang L, Qu G, Jiang YY, Ji C, Zhang HY (2008) Characters of very ancient proteins. Biochem Biophy Res Commun 366:607–611
Mansy SS, Szostak JW (2008) Thermostability of model protocell membranes. Proc Natl Acad Sci USA 105:13351–13355
Mansy SS, Schrum JP, Krishnamurthy M, Tobe S, Treco DA, Szostak JW (2008) Replication of a genetic polymer inside of a model protocell. Nature 454:122–125
Margulis L (1981) Symbiosis in cell evolution: life and its environment on the early earth. W. H. Freeman, San Francisco
Margulis L (1991) Symbiogenesis and symbionticism. In: Margulis L, Fester R (eds) Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. MIT Press, Cambridge, pp 1–14
Martin W, Russell MJ (2003) On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci 358:59–85
Martin W, Russell MJ (2007) On the origin of biochemistry at an alkaline hydrothermal vent. Philos Trans R Soc Lond B Biol Sci 362:1887–1926
Martin W, Baross J, Kelley D, Russell MJ (2008) Hydrothermal vents and the origin of life. Nat Rev Microbiol 6:805–814
Miller S, Krijnse-Locker J (2008) Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6:363–374
Miller SL, Schopf JW, Lazcano A (1997) Oparin’s “origin of Life”: sixty years later. J Mol Evol 44:351–353
Milshteyn D, Damer B, Havig J, Deamer D (2018) Amphiphilic compounds assemble into membranous vesicles in hydrothermal hot spring water but not in sea water. Life 8:11
Moreira D, Lombard J, López-García P (2009) Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol 7:306–311
Mulkidjanian AY, Galperin MY, Koonin EV (2009) Coevolution of primordial membranes and membrane proteins. Trends Biochem Sci 34:206–215
Mulkidjanian AY, Bychkov AY, Dibrova DV, Galperin MY, Koonin EV (2012) Origin of first cells at terrestrial, anoxic geothermal fields. Proc Natl Acad Sci USA 109:E821–E830
Norris V, Raine DJ (1998) A fission-fusion origin for life. Orig Life Evol Biosph 28:523–537
Nunoura K, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, Takai K, Takami H (2011) Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 39:3204–3223
Olasagasti F, Rajamani S (2019) Lipid-assisted polymerization of nucleotides. Life. https://doi.org/10.3390/life9040083
O’Malley MA (2014) Endosymbiosis and its implications for evolutionary theory. Proc Natl Acad Sci USA 112:10270–10277
Oparin AI (1953 [1938]) The origin of life, 2nd edn. Dover, New York [First published by MacMillan.]
Orgel LE (2004) Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol 39:99–123
Pascal R, Pross A (2016) The logic of life. Orig Life Evol Biosph 46:507–513
Pearce BKD, Pudritz RE, Semenov DA, Henning TK (2017) Origin of the RNA world: the fate of nucleobases in warm little ponds. Proc Natl Acad Sci USA 114:11327–11332
Peretó J, López-García P, Moreira D (2004) Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem Sci 29:469–477
Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, Williams LD (2015) History of the ribosome and the origin of translation. Proc Natl Acad Sci USA 112:15396–15401
Philippe H, Forterre P (1999) The rooting of the universal tree of life is not reliable. J Mol Evol 49:509–523
Pohorille A, Deamer D (2009) Self-assembly and function of primitive cell membranes. Res Microbiol 160:449–456
Pohorille A, Wilson MA, Chipot C (2003) Membrane peptides and their role in protobiological evolution. Orig Life Evol Biosph 33:173–197
Pressman A, Blanco C, Chen IA (2015) The RNA world as a model system to study the origin of life. Curr Biol Rev 25:R953–R963
Prosdocimi F, José MV, de Farias ST (2019) The first universal common ancestor (FUCA) as the earliest ancestor of LUCA’s (last UCA) lineage. In: Pontarotti P (ed) Evolution, origin of life, concepts and methods. Springer, Berlin, pp 43–54
Pross A (2011) Toward a general theory of evolution: extending Darwinian theory to inanimate matter. J Syst Chem 2:1
Pross A (2013) The evolutionary origin of biological function and complexity. J Mol Evol 76:185–191
Puigbò P, Wolf YI, Koonin EV (2009) Search for a ‘tree of life’ in the thicket of the phylogenetic forest. J Biol 8:59
Qiao H, Hu N, Bai J, Ren L, Liu Q, Fang L, Wang Z (2017) Encapsulation of nucleic acids into giant unilamellar vesicles by freeze-thaw: a way protocells may form. Orig Life Evol Biosph 47:499–510
Ranea JA, Sillero A, Thornton JM, Orengo CA (2006) Protein superfamily evolution and the last universal common ancestor. J Mol Evol 63:513–525
Richter K, Haslbeck M, Buchner J (2010) The heat shock response: life on the verge of death. Mol Cell 40:253–266
Robertson MP, Joyce GF (2012) The origins of the RNA world. Cold Spring Harb Perspect Biol 4:a003608
Saad NY (2018) A ribonucleopeptide world at the origin of life. J Syst Evol 56:1–13
Sagan L (1967) On the origin of mitosing cells. J Theoret Biol 14:225–274
Saha R, Chen IA (2015) Origin of life: protocells red in tooth and claw. Curr Biol 25:R1175–R1177
Saha R, Pohorille A, Chen IA (2014) Molecular crowding and early evolution. Orig Life Evol Biosph 44:319–324
Saha R, Verbanic S, Chen IA (2018) Lipid vesicles chaperone an encapsulated RNA aptamer. Nat Commun 9:2313
Schrum JP, Zhu TF, Szostak JW (2010) The origins of cellular life. Cold Spring Harb Perspect Biol 2:a002212
Segré D, Ben-Eli D, Deamer DW, Lancet D (2001) The lipid world. Orig Life Evol Biosph 31:119–145
Sengupta S, Higgs PG (2015) Pathways of genetic code evolution in ancient and modern organisms. J Mol Evol 80:229–243
Shah V, de Bouter J, Pauli Q, Tupper AS, Higgs PG (2019) Survival of RNA replicators is much easier in protocells than in surface based, spatial systems. Life 9:65
Shimada H, Yamagishi A (2011) Stability of heterochiral hybrid membrane made of bacterial sn-G3P lipids and archaeal sn-G1P lipids. Biochemistry 50:4114–4120
Smith E, Morowitz HJ (2016) The origin and nature of life on earth. Cambridge University Press, Cambridge
Sojo V (2019) Why the lipid divide? Membrane proteins as drivers of the split between the lipids of the three domains of life. BioEssays 2019(41):1800251
Sojo V, Dessimoz C, Pomiankowski A, Lane N (2016) Membrane proteins are dramatically less conserved than water-soluble proteins across the tree of life. Mol Biol Evol 33:2874–2884
Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG (2015) Complex Archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521:173–179
Spitzer J (2017) Emergence of life on earth: a physicochemical jigsaw puzzle. J Mol Evol 84:1–7
Spitzer J, Poolman B (2009) The role of biomacromolecular crowding, lonic strength and physicochemical gradients in the complexities of life’s emergence. Microbiol Mol Biol Rev 73:371–388
Spitzer J, Poolman B (2016) Complex molecular mixtures under cycling gradients as basis for life’s origins. bioRxiv. https://doi.org/10.1101/050740
Spitzer J, Pielak GJ, Poolman B (2015) Emergence of life: physical chemistry changes the paradigm. Biol Direct 10:33
Staley JT, Fuerst JA (2017) Ancient, highly conserved protein from a LUCA with complex cell biology provide evidence in support of the nuclear compartment commonality (NuCom) hypothesis. Res Microbiol 168:395–412
Storz G, Wolf YI, Ramamurthi KS (2014) Small proteins can no longer be ignored. Annu Rev Biochem 83:753–777
Szathmáry E, Maynard Smith J (1997) From replicators to reproducers: the first major transitions leading to life. J Theor Biol 187:555–571
Szostak JW (2011) An optimal degree of physical and chemical heterogeneity for the origin of life? Philos Trans R Soc B Biol Sci 366:2894–2901
Szostak JW, Bartel DP, Luisi PL (2001) Synthesizing life. Nature 409:387–390
Tagami S, Attwater J, Holliger P (2017) Simple peptides derived from the ribosomal core potentiate RNA polymerase ribozyme function. Nat Chem 9:325–332
Takagi YA, Nguyen DH, Wexler TB, Goldman AD (2020) The Coevolution of cellularity and metabolism following the origin of life. J Mol Evol 88:698–617
Tessera M (2018) Is pre-Darwinian evolution possible? Biol Direct 13:18
Thomas JA, Rana FR (2007) The influence of environmental conditions, lipid composition, and phase behavior on the origin of cell membranes. Orig Life Evol Biosph 37:267–285
Toman J, Flegr J (2017) Stability-based sorting: the forgotten process behind (not only) biological evolution. J Theor Biol 435:29–41
van der Schaar HM, Dorobantu CM, Albulescu L, Strating JRPM, van Kuppeveld FJM (2016) Fat(al) attraction: picornaviruses usurp lipid transfer at membrane contact sites to create replication organelles. Trends Microbiol 24:535–546
van der Veen AG, Ploeh HL (2012) Ubiquitin-like proteins. Annu Rev Biochem 81:323–357
Villanueva L, von Meijenfeldt FAB, Westbye AB, Yadav S, Hopmans EC, Dutilh BE, Sinninghe Damsté JS (2020) Bridging the membrane lipid divide: Bacteria of the FCB group superphylum have the potential to synthesize archaeal ether lipids. ISME J. https://doi.org/10.1038/s41396-020-00772-2
Vetsigian K, Woese C, Goldenfeld N (2006) Collective evolution and the genetic code. Proc Natl Acad Sci USA 103:10696–10701
Vitas M, Dobovišek A (2018) In the beginning was a mutualism-on the origin of translation. Orig Life Evol Biosph 48:223–243
Wächtershäuser G (2003) From pre-cells to Eukarya–a tale of two lipids. Mol Microbiol 47:13–22
Wächtershäuser G (2007) On the chemistry and evolution of the pioneer organism. Chem Biodivers 4:584–602
Wei C, Pohorille A (2011) Permeation of nucleosides through lipid bilayers. J Phys Chem B 115:3681–3688
Wei C, Pohorille A (2013) Permeation of aldoentoses and nucleosides through fatty acid and phospholipid membranes: implication to the origins of life. Astrobiology 13:177–188
Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin MF (2016) The physiology and habitat of the last universal common ancestor. Nat Microbiol 1:16116
White HB (1976) Coenzymes as fossils of an earlier metabolic state. J Mol Evol 7:101–104
Williams TA, Foster PG, Cox CJ, Embley TM (2013) An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504:231–236
Wilson MA, Wei C, Pohorille A (2014) Towards coevolution of membrane proteins and metabolism. Orig Life Evol Biosph 44:357–361
Woese CR (1982) Archaebacteria and cellular origins: an overview. Zbl Bakt Hygi Abr Orig C3:1–17
Woese CR (1998) The universal ancestor. Proc Natl Acad Sci USA 95:6854–6859
Woese CR (2000) Interpreting the universal phylogenetic tree. Proc Natl Acad Sci USA 97:8392–8396
Woese CR (2001) Translation: in retrospect and prospect. RNA 7:1055–1067
Woese CR (2002) On the evolution of cells. Proc Natl Acad Sci USA 99:8742–8747
Woese CR (2004) A new biology for a new century. Microbiol Mol Biol Rev 68:173–186
Woese CR, Fox GE (1977) The concept of cellular evolution. J Mol Evol 10:1–6
Woese CR, Magrum LJ, Fox GE (1978) Archaebacteria. J Mol Evol 11:245–252
Wolf YI, Koonin EV (2007) On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol Direct 2:14. https://doi.org/10.1186/1745-6150-2-14
Wolf YI, Aravind L, Grishin NV, Koonin EV (1999) Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res 9(8):689–710
Wong JT (1975) A coevolution theory of the genetic code. Proc Natl Acad Sci USA 72:1909–1912
Wong JT (1981) Coevolution of genetic code and amino acid biosynthesis. Trends Biochem Sci 6:33–36
Yarus M, Widmann JJ, Knight R (2009) RNA–amino acid binding: a stereochemical era for the genetic code. J Mol Evol 69:406–429
Yarus M (2017) The genetic code and RNA-amino acid affinities. Life 7:13
Zhu TF, Szostak JW (2009) Coupled growth and division of model protocell membranes. J Am Chem Soc 131:5705–5713
Zhu TF, Adamala K, Zhang N, Szostak JW (2012) Photochemically driven redox chemistry induces protocell membrane pearling and division. Proc Natl Acad Sci USA 25:9828–9832
Zimorski V, Ku C, Martin WF, Gould SB (2014) Endosymbiotic theory for organelle origins. Curr Opin Microbiol 22:38–48
Acknowledgements
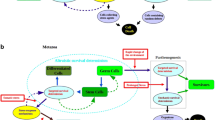
For insightful comments and suggestions, I thank Bruce Damer, David Deamer, Aaron Goldman, Eugene Koonin, four anonymous reviewers, and the editor. Jin He and Tian Yue have provided outstanding research assistance. Special thanks go to Chang-an Liu for drawing the two figures.
Funding
The search has not received any funding.
Author information
Authors and Affiliations
Contributions
ST developed the theory and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no competing interests.
Additional information
Handling editor: Aaron Goldman.
Rights and permissions
About this article
Cite this article
Tang, S. The Origin(s) of Cell(s): Pre-Darwinian Evolution from FUCAs to LUCA. J Mol Evol 89, 427–447 (2021). https://doi.org/10.1007/s00239-021-10014-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-021-10014-4