Abstract
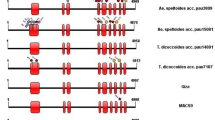
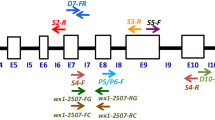
Sweet corn is popular worldwide as vegetable. Though large numbers of sugary1 (su1)-based sweet corn germplasm are available, allelic diversity in su1 gene encoding SU1 isoamylase among diverse maize inbreds has not been analyzed. Here, we characterized the su1 gene in maize and compared with allied species. The entire su1 gene (11,720 bp) was sequenced among six mutant (su1) and five wild (Su1) maize inbreds. Fifteen InDels of 2–45 bp were selected to develop markers for studying allelic diversity in su1 gene among 19 mutant- (su1) and 29 wild-type (Su1) inbreds. PIC ranged from 0.15 (SU-InDel7) to 0.37 (SU-InDel13). Major allele frequency varied from 0.52 to 0.90, while gene diversity ranged from 0.16 to 0.49. Phylogenetic tree categorized 48 maize inbreds in two clusters each for wild- type (Su1) and mutant (su1) types. 44 haplotypes of su1 were observed, with three haplotypes (Hap6, Hap22 and Hap29) sharing more than one genotype. Further, comparisons were made with 23 orthologues of su1 from 16 grasses and Arabidopsis. Maize possessed 15–19 exons in su1, while it was 11–24 exons among orthologues. Introns among the orthologues were longer (77–2206 bp) than maize (859–1718 bp). SU1 protein of maize and orthologues had conserved α-amylase and CBM_48 domains. The study also provided physicochemical properties and secondary structure of SU1 protein in maize and its orthologues. Phylogenetic analysis showed closer relationship of maize SU1 protein with P. hallii, S. bicolor and E. tef than Triticum sp. and Oryza sp. The study showed that presence of high allelic diversity in su1 gene which can be utilized in the sweet corn breeding program. This is the first report of comprehensive characterization of su1 gene and its allelic forms in diverse maize and related orthologues.






Similar content being viewed by others
Data availability
The data on su1 gene sequences generated in the present study have been submitted to GenBank at NCBI, with the submission ID 2468400, dated: 04-06-2021.
References
Amemura A, Chakraborty R, Fujita M, Noumi T, Futai M (1988) Cloning and nucleotide sequence of the isoamylase gene from Pseudomonas amyloderamosa SB-15. J Biol Chem 263:9271–9275
Baseggio M, Murray M, Magallanes-Lundback M, Kaczmar N, Chamness J, Buckler ES, Smith ME, DellaPenna D, Tracy WF, Gore MA (2020) Natural variation for carotenoids in fresh kernels is controlled by uncommon variants in sweet corn. Plant Genome 13:e20008. https://doi.org/10.1002/tpg2.20008
Baum BR, Johnson DA (2008) Molecular confirmation of the genomic constitution of Douglasdeweya (Triticeae: Poaceae): demonstration of the utility of the 5S rDNA sequence as a tool for haplome identification. Mol Genet Genom 279:621–628
Baveja A, Muthusamy V, Panda KK, Zunjare RU, Das AK, Chhabra R, Mishra SJ, Mehta BK, Saha S, Hossain F (2021) Development of multinutrient-rich sweet corn hybrids through genomics-assisted selection of shrunken2, opaque2, lcyE and crtRB1 genes. J Appl Genet. https://doi.org/10.1007/s13353-021-00633-4
Branden C, Tooze J (1999) Introduction to protein structure. Garland Publishing, Taylor and Francis Group, New York
Burton RA, Jenner H, Carrangis L, Fahy B, Fincher GB, Hylton C, Laurie DA, Parker M, Waite D, van Wegen S, Verhoeven T, Denyer K (2002) Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J 31:97–112. https://doi.org/10.1046/j.1365-313x.2002.01339.x
Buske FA, Bodén M, Bauer DC, Bailey TL (2010) Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics 26:860–866. https://doi.org/10.1093/bioinformatics/btq049
Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD (2013) An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinform 14:1–8. https://doi.org/10.1186/1471-2105-14-S2-S4
Cheng K, Zhang F, Sun F, Chen H, Zhang YP (2015) Doubling power output of starch biobattery treated by the most thermostable isoamylase from an archaeon Sulfolobus tokodaii. Sci Rep 5:1–10. https://doi.org/10.1038/srep13184
Chhabra R, Hossain F, Muthusamy V, Baveja A, Mehta BK, Zunjare RU (2019) Development and validation of breeder-friendly functional markers of sugary1 gene encoding starch-debranching enzyme affecting kernel sweetness in maize (Zea mays). Crop Pasture Sci 70:868–875. https://doi.org/10.1071/CP19298
Chhabra R, Hossain F, Muthusamy V, Baveja A, Mehta BK, Zunjare RU (2020) Development and validation of gene-based markers for shrunken2-Reference allele and their utilization in marker-assisted sweet corn (Zea mays Sachharata) breeding programme. Plant Breed. https://doi.org/10.1111/pbr.12872
Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J (2013) Haplotype estimation using sequencing reads. Am J Hum Genet 93:687–696. https://doi.org/10.1016/j.ajhg.2013.09.002
Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41:815–830
Dellaporta SL, Wood J, Hicks JB (1985) Maize DNA miniprep. In: Malberg J, Messing, Sussex I (eds) Plant molecular biology. Cold Spring Harbor, New York, p 363
Dinges JR, Colleoni C, Myers AM, James MG (2001) Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol 125:1406–1418. https://doi.org/10.1104/pp.125.3.1406
FAOSTAT (2019) Trade data: crops and livestock products. http://www.fao.org/faostat/en/#data/TP
Feng ZL, Liu J, Fu FL, Li WC (2008) Molecular mechanism of sweet and waxy in maize. Int J Plant Breed Genet 2:93–100. https://doi.org/10.3923/ijpbg.2008.93.100
Fujita N, Kubo A, Francisco PB Jr, Nakakita M, Harada K, Minaka N, Nakamura Y (1999) Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta 208:283–293
Gomes I, Gomes J, Steiner W (2003) Highly thermostable amylase and pullulanase of the extreme thermophilic eubacterium Rhodothermus marinus: production and partial characterization. Bioresour Technol 90:207–214. https://doi.org/10.1016/s0960-8524(03)00110-x
Hossain F, Nepolean T, Vishwakarma AK, Panday N, Prasanna BM, Gupta HS (2015) Mapping and validation of microsatellite markers linked to sugary1 and shrunken2 genes in maize. J Plant Biochem Biotechnol. https://doi.org/10.1007/s13562-013-0245-3
Hung NT, Huyen TN, Loc NV, Cuong BM (2012) The application of SSR molecular indicator to assess the purity and genetic diversity of waxy corn inbred lines. J ISSASS 18:45–54
Hussain H, Mant A, Seale R, Zeeman S, Hinchliffe E, Edwards A, Hylton C, Bornemann S, Smith AM, Martin C, Bustos R (2003) Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell 15:133–149. https://doi.org/10.1105/tpc.006635
James MG, Robertson DS, Myers AM (1995) Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7:417–429. https://doi.org/10.1105/tpc.7.4.417
Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, Humphrey J, Kerhornou A, Khobova J, Aranganathan NK, Langridge N, Lowy E, McDowall MD, Maheswari U, Nuhn M, Staines DM (2016) Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res 44(D1):D574-580. https://doi.org/10.1093/nar/gkv1209
Krieger F, Moglich A, Kiefhaber T (2004) Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J Am Chem Soc 127:3346–3352. https://doi.org/10.1021/ja042798i
Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y (1999) The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol 121:399–410. https://doi.org/10.1104/pp.121.2.399
Kubo A, Rahman S, Utsumi Y, Li Z, Mukai Y, Yamamoto M, Ugaki M, Harada K, Satoh H, Konik-Rose C, Morell M, Nakamura Y (2005) Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase1 gene supports a direct role for isoamylase1 in amylopectin biosynthesis. Plant Physiol 137:43–56. https://doi.org/10.1104/pp.104.051359
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. https://doi.org/10.1016/0022-2836(82)90515-0
Law MJ, Linde ME, Chambers EJ, Oubridge C, Katsamba PS, Nilsson L, Haworth IS, Laird-Offringa IA (2006) The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res 34:275–285. https://doi.org/10.1093/nar/gkj436
Lertrat K, Pulam T (2007) Breeding for increased sweetness in sweet corn. Int J Plant Breed 1:27–30
Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz239
Lin Q, Huang B, Zhang M, Zhang X, Rivenbark J, Lappe RL, James MG, Myers AM, Hennen-Bierwagen TA (2012) Functional interactions between starch synthase III and isoamylase-type starch-debranching enzyme in maize endosperm. Plant Physiol 158:679–692. https://doi.org/10.1104/pp.111.189704
Liu K, Muse SV (2005) PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21:2128–2129
Mayer M, Hölker AC, González-Segovia E, Bauer E, Presterl T, Ouzunova M, Melchinger AE, Schön CC (2020) Discovery of beneficial haplotypes for complex traits in maize landraces. Nat Commun 11:4954. https://doi.org/10.1038/s41467-020-18683-3
Mehta B, Hossain F, Muthusamy V, Baveja A, Zunjare R, Jha SK, Gupta HS (2017) Microsatellite-based genetic diversity analyses of sugary1-, shrunken2- and double mutant- sweet corn inbreds for their utilization in breeding programme. Physiol Mol Biol Plants 23:411–420. https://doi.org/10.1007/s12298-017-0431-1
Mehta BK, Muthusamy V, Zunjare RU, Baveja A, Chauhan HS, Chhabra R, Hossain F (2020) Biofortification of sweet corn hybrids for provitamin-A, lysine and tryptophan using molecular breeding. J Cereal Sci 96:103093. https://doi.org/10.1016/j.jcs.2020.103093
Neuffer MG, Coe EH, Wessler SR (1997) Mutants of maize. Harbor Laboratory Press, Cold Spring
Odibo FJC, Okafor N, Tom MU, Oyeka CA (1992) Purification and characterization of a β-Amylase of Hendersonula toruloidea. Starch-Starke 44:192–195. https://doi.org/10.1002/star.19920440508
Olempska-Beer ZS, Merker RI, Ditto MD, DiNovi MJ (2007) Food-processing enzymes from recombinant microorganisms—a review. Regul Toxicol Pharmacol 45:144–158. https://doi.org/10.1016/j.yrtph.2006.05.001
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. CRC Press, New York, pp 43–76
Rahman A, Wong KS, Jane JL, Myers AM, James MG (1998) Characterization of SU1 isoamylase, a determinant of storage starch structure in maize. Plant Physiol 117:425–435. https://doi.org/10.1104/pp.117.2.425
Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56
Rogers S, Wells R, Rechsteiner M (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234:364–368
Shin JH, Kwon SJ, Lee JK, Min HK, Kim NS (2006) Genetic diversity of maize kernel starch-synthesis genes with SNAPs. Genome 49:1287–1296. https://doi.org/10.1139/g06-116
Sim L, Beeren SR, Findinier J, Dauvillée D, Ball SG, Henriksen A, Palcic MM (2014) Crystal structure of the Chlamydomonas starch debranching enzyme isoamylase ISA1 reveals insights into the mechanism of branch trimming and complex assembly. J Biol Chem 289:22991–23003. https://doi.org/10.1074/jbc.m114.565044
Solovyev V, Kosarev P, Seledsov I, Vorobyev D (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 7:1–2. https://doi.org/10.1186/gb-2006-7-s1-s10
Sood S, Flint-Garcia S, Willcox MC, Holland JB (2014) Mining natural variation for maize improvement: selection on Phenotypes and Genes. In: Tuberosa R, Graner A, Frison E (eds) Genomics of plant genetic resources. Springer, Amsterdam, pp 615–649
Tracy WF, Whitt SR, Buckler ES (2006) Recurrent mutation and genome evolution: example of Sugary1 and the origin of sweet maize. Crop Sci 46:S49-54. https://doi.org/10.2135/cropsci2006-03-0149tpg
Utsumi Y, Nakamura Y (2006) Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the Isoamylase1-isoamylase2 hetro-oligomer from rice endosperm. Planta 225:75–87
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, De Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296-303. https://doi.org/10.1093/nar/gky427
Yang R, Yan Z, Wang Q, Li X, Feng F (2018) Marker-assisted backcrossing of lcyE for enhancement of proA in sweet corn. Euphytica 214:1–2. https://doi.org/10.1007/s10681-018-2212-5
Zhang MZ, Fang JH, Yan X, Liu J, Bao J-S, Fransson G, Andersson R, Jansson C, Aman P, Sun C (2012) Molecular insights into how a deficiency of amylose affects carbon allocation-carbohydrate and oil analyses and gene expression profiling in the seeds of a rice waxy mutant. BMC Plant Biol 12:1–18
Acknowledgements
Financial help received from under “Women Scientist-A Scheme” (File No. WOS-A/LS-306/2018), Department of Science and Technology, Govt. of India, and “Young Scientists' Scheme” [Sanction No. YSS/2015/001029], Science and Engineering Research Board (SERB) is thankfully acknowledged. We thank ICAR-IARI, New Delhi for providing field and lab facilities for the conduct of the experiment.
Author information
Authors and Affiliations
Contributions
Conduct of the experiment: RC, Designing of markers: MV, In-silico data generation: NG, Marker assays: AK, NRP, Development and maintenance of mutant inbreds: MV and NRP Allelic diversity analysis: RUZ, Drafting of manuscript: RC and FH, Design of experiment: FH and RC.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chhabra, R., Muthusamy, V., Gain, N. et al. Allelic variation in sugary1 gene affecting kernel sweetness among diverse-mutant and -wild-type maize inbreds. Mol Genet Genomics 296, 1085–1102 (2021). https://doi.org/10.1007/s00438-021-01807-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-021-01807-9