Abstract
NMR chemical shifts (CSs) are delicate reporters of local protein structure, and recent advances in random coil CS (RCCS) prediction and interpretation now offer the compelling prospect of inferring small populations of structure from small deviations from RCCSs. Here, we present CheSPI, a simple and efficient method that provides unbiased and sensitive aggregate measures of local structure and disorder. It is demonstrated that CheSPI can predict even very small amounts of residual structure and robustly delineate subtle differences into four structural classes for intrinsically disordered proteins. For structured regions and proteins, CheSPI provides predictions for up to eight structural classes, which coincide with the well-known DSSP classification. The program is freely available, and can either be invoked from URL www.protein-nmr.org as a web implementation, or run locally from command line as a python program. CheSPI generates comprehensive numeric and graphical output for intuitive annotation and visualization of protein structures. A number of examples are provided.




taken from a 2D-color-scale (see Fig. S3) based on the position in Ramachandran plot of pairs of ϕ and Ψ backbone torsion angles using trigonometric averages of the ensemble values (see Eq. 16, Methods). With this scale, backbone angles in the helical domain of the Ramachandran map appear in red as before, and extended β-sheet-like conformations have blue colors. Furthermore, left-twisted β-strands as well as fragments with PPII structure appear with cyan colors, whereas conformations with positive ϕ have yellow and green colors, and finally, other conformation referred to elsewhere as “forbidden” in Ramachandran space are shown in black. Transparency is added to the bars using the above local angular order parameters as the “alpha value”. See also Figure S1







Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. Python code for CheSPI is available for download at GitHub: https://github.com/protein-nmr.
References
Adzhubei AA, Sternberg MJ, Makarov AA (2013) Polyproline-II helix in proteins: structure and function. J Mol Biol 425:2100–2132
Banci L et al (2002) The solution structure of reduced dimeric copper zinc superoxide dismutase. Eur J Biochem 269:1905–1915
Barghorn S, Davies P, Mandelkow E (2004) Tau paired helical filaments from Alzheimer’s disease brain and assembled in vitro are based on beta-structure in the core domain. Biochemistry 43:1694–1703
Berjanskii MV, Wishart DS (2007) The RCI server: rapid and accurate calculation of protein flexibility using chemical shifts. Nucleic Acids Res 35:W531–W537
Bernadó P et al (2005) A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc Natl Acad Sci USA 102:17002–17007
Berriman J et al (2003) Tau filaments from human brain and from in vitro assembly of recombinant protein show cross-beta structure. Proc Natl Acad Sci USA 100:9034–9038
Bradley W, Robert P (2013) Multivariate analysis in metabolomics. Curr Metabol 1:92–107
Braun D, Wider G, Wuethrich K (1994) Sequence-corrected 15N “random coil” chemical shifts. J Am Chem Soc 116:8466–8469
Brutscher B et al (2015) NMR methods for the study of instrinsically disordered proteins structure, dynamics, and interactions: general overview and practical guidelines. Adv Exp Med Biol 870:49–122
Bunney TD et al (2006) Structural and mechanistic insights into ras association domains of phospholipase C epsilon. Mol Cell 21:495–507
Bylesjö M et al (2006) OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemom 20:341–351
Camilloni C, De Simone A, Vranken WF, Vendruscolo M (2012) Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry 51:2224–2231
Camilloni C, Cavalli A, Vendruscolo M (2013) Replica-averaged metadynamics. J Chem Theory Comput 9:5610–5617
Cavalli A, Salvatella X, Dobson CM, Vendruscolo M (2007) Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci 104:9615
Cleveland DW, Hwo SY, Kirschner MW (1977) Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J Mol Biol 116:227–247
Daebel V et al (2012) β-Sheet core of Tau paired helical filaments revealed by solid-state NMR. J Am Chem Soc 134:13982–13989
Dass R, Mulder FAA, Nielsen JT (2020) ODiNPred: comprehensive prediction of protein order and disorder. Sci Rep 10:14780
Davidson WS, Jonas A, Clayton DF, George JM (1998) Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem 273:9443–9449
De Simone A et al (2009) Accurate random coil chemical shifts from an analysis of loop regions in native states of proteins. J Am Chem Soc 131:16332–16333
Eghbalnia HR et al (2005) Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J Biomol NMR 32:71–81
Eliezer D et al (2005) Residual structure in the repeat domain of tau: echoes of microtubule binding and paired helical filament formation. Biochemistry 44:1026–1036
Felli IC, Pierattelli R (2012) Recent progress in NMR spectroscopy: toward the study of intrinsically disordered proteins of increasing size and complexity. IUBMB Life 64:473–481
Fitzpatrick AWP et al (2017) Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547:185–190
Graham LA, Davies PL (2005) Glycine-rich antifreeze proteins from snow fleas. Science 310:461
Hafsa NE, Arndt D, Wishart DS (2015) CSI 3.0: a web server for identifying secondary and super-secondary structure in proteins using NMR chemical shifts. Nucleic Acids Res 43:W370–W377
Hyberts SG, Goldberg MS, Havel TF, Wagner G (1992) The solution structure of eglin c based on measurements of many NOEs and coupling constants and its comparison with X-ray structures. Protein Sci 1:736–751
Jensen PH et al (1998) Binding of alpha-synuclein to brain vesicles is abolished by familial Parkinson’s disease mutation. J Biol Chem 273:26292–26294
Jensen MR, Salmon L, Nodet G, Blackledge M (2010) Defining conformational ensembles of intrinsically disordered and partially folded proteins directly from chemical shifts. J Am Chem Soc 132:1270–1272
Jha AK, Colubri A, Freed KF, Sosnick TR (2005) Statistical coil model of the unfolded state: resolving the reconciliation problem. Proc Natl Acad Sci USA 102:13099
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Källberg M et al (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1522
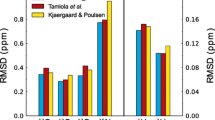
Kjaergaard M, Poulsen FM (2012) Disordered proteins studied by chemical shifts. Prog Nucl Magn Reson Spectrosc 60:42–51
Kjaergaard M, Brander S, Poulsen FM (2011) Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J Biomol NMR 49:139–149
Kukic P et al (2019) The free energy landscape of the oncogene protein E7 of human papillomavirus type 16 reveals a complex interplay between ordered and disordered regions. Sci Rep 9:5822. https://doi.org/10.1038/s41598-019-41925-4
Labudde D, Leitner D, Krüger M, Oschkinat H (2003) Prediction algorithm for amino acid types with their secondary structure in proteins (PLATON) using chemical shifts. J Biomol NMR 25:41–53
Lee JH et al (2015) Heterogeneous binding of the SH3 client protein to the DnaK molecular chaperone. Proc Natl Acad Sci USA 112:E4206-4215
Makowska J et al (2006) Polyproline II conformation is one of many local conformational states and is not an overall conformation of unfolded peptides and proteins. Proc Natl Acad Sci USA 103:1744
Marsh JA, Forman-Kay JD (2009) Structure and disorder in an unfolded state under nondenaturing conditions from ensemble models consistent with a large number of experimental restraints. J Mol Biol 391:359–374
Marsh JA, Forman-Kay JD (2012) Ensemble modeling of protein disordered states: experimental restraint contributions and validation. Proteins 80:556–572
Marsh JA, Singh VK, Jia Z, Forman-Kay JD (2006) Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci 15:2795–2804
Mielke SP, Krishnan VV (2009) Characterization of protein secondary structure from NMR chemical shifts. Prog Nucl Magn Reson Spectrosc 54:141–165
Milani P, Gagliardi S, Cova E, Cereda C (2011) SOD1 Transcriptional and posttranscriptional regulation and its potential implications in ALS. Neurol Res Int 2011:458427
Mukrasch MD et al (2007) Highly populated turn conformations in natively unfolded tau protein identified from residual dipolar couplings and molecular simulation. J Am Chem Soc 129:5235–5243
Neal S, Nip AM, Zhang HY, Wishart DS (2003) Rapid and accurate calculation of protein H-1, C-13 and N-15 chemical shifts. J Biomol NMR 26:215–240
Nielsen JT, Mulder FAA (2016) There is diversity in disorder—“In all chaos there is a cosmos, in all disorder a secret order.” Front Mol Biosci. https://doi.org/10.3389/fmolb.2016.00004
Nielsen JT, Mulder FAA (2018) POTENCI: prediction of temperature, neighbor and pH-corrected chemical shifts for intrinsically disordered proteins. J Biomol NMR 70:141–165
Nielsen JT, Mulder FAA (2019) Quality and bias of protein disorder predictors. Sci Rep 9:5137
Nielsen JT, Mulder FAA (2020) Quantitative protein disorder assessment Using NMR chemical shifts. In: Kragelund BB, Skriver K (eds) Intrinsically Disordered proteins: methods and protocols. Springer, New York, pp 303–317
Nielsen JT, Eghbalnia HR, Nielsen NC (2012) Chemical shift prediction for protein structure calculation and quality assessment using an optimally parameterized force field. Prog Nucl Magn Reson Spectrosc 60:1–28
Ozenne V et al (2012) Mapping the potential energy landscape of intrinsically disordered proteins at amino acid resolution. J Am Chem Soc 134:15138–15148
Pal L, Chakrabarti P, Basu G (2003) Sequence and structure patterns in proteins from an analysis of the shortest helices: implications for helix nucleation. J Mol Biol 326:273–291
Pentelute BL et al (2008) X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers. J Am Chem Soc 130:9695–9701
Rakhit R, Chakrabartty A (2006) Structure, folding, and misfolding of Cu, Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochimica et Biophysica Acta Mol Basis Dis 1762:1025–1037
Rao JN, Dua V, Ulmer TS (2008) Characterization of alpha-synuclein interactions with selected aggregation-inhibiting small molecules. Biochemistry 47:4651–4656
Rao JN, Kim YE, Park LS, Ulmer TS (2009) Effect of pseudorepeat rearrangement on alpha-synuclein misfolding, vesicle binding, and micelle binding. J Mol Biol 390:516–529
Rao JN et al (2010) A Combinatorial NMR and EPR approach for evaluating the structural ensemble of partially folded proteins. J Am Chem Soc 132:8657–8668
Richardson JS, Richardson DC (2002) Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA 99:2754–2759
Robberecht W et al (1994) Cu/Zn superoxide dismutase activity in familial and sporadic amyotrophic lateral sclerosis. J Neurochem 62:384–387
Robustelli P, Stafford KA, Palmer AG (2012) Interpreting protein structural Dynamics from NMR chemical shifts. J Am Chem Soc 134:6365–6374
Rosen DR et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62
Schwarzinger S et al (2001) Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc 123:2970–2978
Shapovalov M, Vucetic S, Dunbrack RL Jr (2019) A new clustering and nomenclature for beta turns derived from high-resolution protein structures. PLoS Comput Biol 15:e1006844–e1006844
Shen Y, Bax A (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR 56:227–241
Shen Y, Bax A (2015) Protein structural information derived from nmr chemical shift with the neural network program TALOS-N. Methods Mol Biol 1260:17–32
Shi Z, Chen K, Liu Z, Kallenbach NR (2006) Conformation of the backbone in unfolded proteins. Chem Rev 106:1877–1897
Singleton AB et al (2003) [alpha]-synuclein locus triplication causes Parkinson’s disease. Science 302:841
Sirangelo I, Iannuzzi C (2017) The role of metal binding in the amyotrophic lateral sclerosis-related aggregation of copper-zinc superoxide dismutase. Molecules 22:1429
Sormanni P et al (2017) Simultaneous quantification of protein order and disorder. Nat Chem Biol 13:339–342
Stefanis L (2012) α-Synuclein in Parkinson’s disease. Cold Spring Harb Perspect Med 2:a009399–a009399
Sterckx YGJ et al (2014) Small-angle X-Ray scattering- and nuclear magnetic resonance-derived conformational ensemble of the highly flexible antitoxin PaaA2. Structure 22:854–865
Tamiola K, Mulder FAA (2012) Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem Soc Trans 40:1014–1020
Tamiola K, Acar B, Mulder FAA (2010) Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc 132:18000–18003
Teilum K et al (2009) Transient structural distortion of metal-free Cu/Zn superoxide dismutase triggers aberrant oligomerization. Proc Natl Acad Sci USA 106:18273–18278
Tofaris GK, Spillantini MG (2005) Alpha-synuclein dysfunction in Lewy body diseases. Mov Disord 20(Suppl 12):S37-44
Tompa P (2009) Structure and function of intrinsically disordered proteins. Chapman and Hall/CRC, London
Trygg J, Wold S (2002) Orthogonal projections to latent structures (O-PLS). J Chemom 16:119–128
Uversky VN, Longhi S (2010) Instrumental analysis of intrinsically disordered proteins: assessing structure and conformation. Wiley, Hoboken
Varadi M et al (2014) pE-DB: a database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucleic Acids Res 42:D326–D335
Varadi M, Vranken W, Guharoy M, Tompa P (2015) Computational approaches for inferring the functions of intrinsically disordered proteins. Front Mol Biosci. https://doi.org/10.3389/fmolb.2015.00045
von Bergen M et al (2000) Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc Natl Acad Sci USA 97:5129–5134
Wang C-C, Chen J-H, Lai W-C, Chuang W-J (2007) 2DCSi: identification of protein secondary structure and redox state using 2D cluster analysis of NMR chemical shifts. J Biomol NMR 38:57–63
Wang Z, Zhao F, Peng J, Xu J (2011) Protein 8-class secondary structure prediction using conditional neural fields. Proteomics 11:3786–3792
Wishart DS, Case DA (2001) Use of chemical shifts in macromolecular structure determination. Resonance Biologica Macromol C 338:3–34
Wishart DS, Sykes BD (1994a) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4:171–180
Wishart DS, Sykes BD (1994b) Chemical-shifts as a tool for structure determination. Methods Enzymol 239:363–392
Wishart DS, Sykes BD, Richards FM (1992) The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31:1647–1651
Wishart DS et al (1995) 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR 5:67–81
Worley B, Powers R (2016) PCA as a practical indicator of OPLS-DA model reliability. Curr Metabol 4:97–103
Wu Z, Li D, Meng J, Wang H (2010) Introduction to SIMCAP and its application. In: Esposito Vinzi V, Chin WW, Henseler J, Wang H (eds) Handbook of partial least squares concepts methods and applications. Springer, Berlin, pp 757–774
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nielsen, J.T., Mulder, F.A.A. CheSPI: chemical shift secondary structure population inference. J Biomol NMR 75, 273–291 (2021). https://doi.org/10.1007/s10858-021-00374-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-021-00374-w