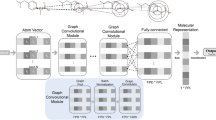
Abstract
Deep neural networks are effective in learning directly from low-level encoded data without the need of feature extraction. This paper shows how QSAR models can be constructed from 2D molecular graphs without computing chemical descriptors. Two graph convolutional neural network-based models are presented with and without a Bayesian estimation of the prediction uncertainty. The property under investigation is mutagenicity: Models developed here predict the output of the Ames test. These models take the SMILES representation of the molecules as input to produce molecular graphs in terms of adjacency matrices and subsequently use attention mechanisms to weight the role of their subgraphs in producing the output. The results positively compare with current state-of-the-art models. Furthermore, our proposed model interpretation can be enhanced by the automatic extraction of the substructures most important in driving the prediction, as well as by uncertainty estimations.
Graphic abstract








Similar content being viewed by others
References
LeCun Y, Bengio Y (1995) Convolutional networks for images, speech, and time series. In Arbib MA (ed) The handbook of brain theory and neural networks, vol. 3361(10)
Mayr A, Klambauer G, Unterthiner T, Hochreiter S (2016) DeepTox: toxicity prediction using deep learning. Front Environ Sci 3:80. https://doi.org/10.3389/fenvs.2015.00080
Gomez-Bombarelli R, Wei JN, Duvenaud D, Hernandez-Lobato JM, Sanchez-Lengeling B, Sheberla D, Aguilera-Iparraguirre J, Hirzel TD, Adams RP, Aspuru-Guzik A (2018) Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent Sci 4:268–276. https://doi.org/10.1021/acscentsci.7b00572
Putin E, Asadulaev A, Ivanenkov Y, Aladinskiy W, Sanchez-Lengeling B, Aspuru-Guzik A, Zhavoronkov A (2018) Reinforced adversarial neural computer for de novo molecular design. J Chem Inf Model 58:1194–1204. https://doi.org/10.1021/acs.jcim.7b00690
Segler MH, Preuss M, Waller MP (2018) Planning chemical syntheses with deep neural networks and symbolic. AI. Nature 555:604. https://doi.org/10.1038/nature25978
Zhou Z, Li X, Zare RN (2017) Optimizing chemical reactions with deep reinforcement learning. ACS Cent Sci 3:1337–1344. https://doi.org/10.1021/acscentsci.7b00492
Winkler DA, Le TC (2017) Performance of deep and shallow neural networks, the universal approximation theorem, activity cliffs, and QSAR. Mol Inform 36(1–2):1600118
Segler MH, Kogej T, Tyrchan C, Waller MP (2017) Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent Sci 4:120–131. https://doi.org/10.1021/acscentsci.7b00512
Smith JS, Isayev O, Roitberg AE (2017) ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem Sci 8:3192–3203. https://doi.org/10.1039/C6SC05720A
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444. https://doi.org/10.1038/nature14539
Winter R, Montanari F, Noé F (2019) Clevert D-A (2019) Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations. Chem Sci 10:1692. https://doi.org/10.1039/c8sc04175j
Bengio Y, Courville A, Vincent P (2013) Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 35(8):1798–1828. https://doi.org/10.1109/TPAMI.2013.50
Goh GB, Siegel C, Vishnu A, Hodas N, Baker N (2018) How much chemistry does a deep neural network need to know to make accurate predictions? In 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), p 1340–1349
Kearnes S, McCloskey K, Berndl M, Pande V, Riley P (2016) Molecular graph convolutions: moving beyond fingerprints. J Comput Aided Mol Des 30(8):595–608. https://doi.org/10.1007/s10822-016-9938-8
Zhou J, Cui G, Zhang Z, Yang C, Liu Z, Wang L, Li C, Sun M (2019) Graph Neural Networks: a review of methods and applications. AI Open 1. https://doi.org/10.1016/j.aiopen.2021.01.001
Roy K (ed) (2017) Advances in QSAR modeling: applications in pharmaceutical chemical food agricultural and environmental sciences. Springer International Publishing, Switzerland
Ryu S, Kwon Y, Kim WY (2019) A Bayesian graph convolutional network for reliable prediction of molecular properties with uncertainty quantification. Chem Sci 10:8438–8446. https://doi.org/10.1039/C9SC01992H
Benigni R, Bossa C (2008) Structure alerts for carcinogenicity, and the Salmonella assay system: a novel insight through the chemical relational databases technology. Mutat Res 659(3):248–261. https://doi.org/10.1016/j.mrrev.2008.05.003
Gini G, Ferrari T, Cattaneo D, Bakhtyari NG, Manganaro A, Benfenati E (2013) Automatic knowledge extraction from chemical structures: the case of mutagenicity prediction. SAR QSAR Environ Res 24(5):365–383. https://doi.org/10.1080/1062936X.2013.773376
Benfenati E, Manganaro A, Gini G (2013) VEGA-QSAR: AI inside a platform for predictive toxicology, Workshop Popularize Artificial Intelligence (PAI) 2013 Torino, http://ceur-ws.org/Vol-1107/
Miller EC (1981) Miller J A (1981) Searches for ultimate chemical carcinogens and their reactions with cellular macromolecules. Cancer 47:2327–2345. https://doi.org/10.1002/1097-0142(19810515)47:10%3c2327::aid-cncr2820471003%3e3.0.co;2-z
Martin YC, Kofron JL, Traphagen LM (2002) Do structurally similar molecules have similar biological activity? J Med Chem 45(19):4350–4358. https://doi.org/10.1021/jm020155c
Hansen K, Mika S, Schroeter T, Sutter A, ter Laak A, Steger-Hartmann T, Heinrich N, Müller K (2009) Benchmark data set for in silico prediction of Ames mutagenicity. J Chem Inf Model 49(9):2077–2081. https://doi.org/10.1021/ci900161g
Kazius J, McGuire R, Bursi R (2005) Derivation and validation of toxicophores for mutagenicity prediction. J Med Chem 48:312–320. https://doi.org/10.1021/jm040835a
Honma M, Kitazawa A, Cayley A, Williams RV, Barber C, Hanser T, Saiakhov R, Chakravarti S, Myatt GJ, Cross KP, Benfenati E, Raitano G, Mekenyan O, Petkov P, Bossa C, Benigni R, Battistelli CL, Giuliani A, Tcheremenskaia O, Rathman J (2019) Improvement of quantitative structure-activity relationship (QSAR) tools for predicting Ames mutagenicity: outcomes of the Ames/QSAR International Challenge Project. Mutagenesis 34(1):3–16. https://doi.org/10.1093/mutage/gey031
Gini G, Katrizky A (Eds.) (1999) Predictive toxicology of chemicals: experiences and impact of AI tools, papers from the AAAI Spring Symposium on Predictive toxicology SS-99-01. AAAI Press, Menlo Park, CA
An G (1996) The effects of adding noise during backpropagation training on a generalization performance. Neural Comput 8:643–674. https://doi.org/10.1162/neco.1996.8.3.643
Weininger M, Weininger A, Weininger JL (1989) SMILES. 2. Algorithm for generation of unique SMILES notation. J Chem Inf Model 29:97–101. https://doi.org/10.1021/ci00062a008
Wu Z, Pan S, Chen F, Long G, Zhang C, Yu PS (2020) A comprehensive survey on Graph Neural Networks. IEEE Trans Neural Netw Learn Syst. https://doi.org/10.1109/TNNLS.2020.2978386
Kipf T N, Welling M (2017) Semi-supervised classification with graph convolutional networks. Proceedings International Conference on Learning Representations (ICLR 2017). https://openreview.net/pdf?id=SJU4ayYgl
Xiong Z, Wang D, Liu X, Zhong F, Wan X, Li X, Li Z, Luo X, Chen K, Jiang H, Zheng M (2020) Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J Med Chem 63(16):8749–8760. https://doi.org/10.1021/acs.jmedchem.9b00959
Mnih V, Heess N, Graves A, Kavukcuoglu K (2014) Recurrent Models of Visual Attention. In Proceedings of NIPS. p 2204–2212
Lee JB, Rossi RA, Kim S, Ahmed NK, Koh E (2019) Attention models in graphs: a survey. ACM Trans Knowl Discov Data. https://doi.org/10.1145/3363574
Velickovic P, Cucurull G, Casanova A, Romero A, Lio P, Bengio Y (2017) Graph attention networks. In Proceedings ICLR. https://doi.org/10.17863/CAM.48429
Gal Y, Ghahramani Z (2016) Dropout as a Bayesian approximation: representing model uncertainty in deep learning. In Proceedings of the 33rd International conference on machine learning, PMLR 48: 1050–1059
Gal Y, Hron J (2017) Concrete dropout. In Proceedings 31st International Conference on neural information processing systems, December, p 3584–3593
Der Kiureghian A, Ditlevsen O (2009) Aleatory or epistemic? does it matter? Struct Saf 31:105–112. https://doi.org/10.1016/j.strusafe.2008.06.020
Kendall A, Gal Y (2017) What uncertainties do we need in Bayesian deep learning for computer vision? Advances in neural information processing systems. 5574– 5584
Ames BN (1984) The detection of environmental mutagens and potential. Cancer 53:2030–2040. https://doi.org/10.1002/1097-0142(19840515)53:10%3c2034::aid-cncr2820531005%3e3.0.co;2-s
Branco P, Torgo L, Ribeiro RP (2015) A survey of predictive modeling under imbalanced distributions. arXiv:1505.01658v2 [cs.LG]
Piegorsch WW, Zeiger E (1991) Measuring intra-assay agreement for the Ames salmonella assay. In: Hotorn L (ed) Statistical methods in toxicology. Springer-Verlag, Berlin, pp 35–41
Zur RM, Jiang Y, Pesce LL, Drukker K (2009) Noise injection for training artificial neural networks: a comparison with weight decay and early stopping. Med Phys 36(10):4810–4818. https://doi.org/10.1118/1.3213517
Polishchuk PG (2017) Interpretation of QSAR models: past, present and future. J Chem Inf Model 57(11):2618–2639. https://doi.org/10.1021/acs.jcim.7b00274
Benigni R, Bossa C, Jeliazkova N, Netzeva T, Worth A (2008) The Benigni/Bossa rulebase for mutagenicity and carcinogenicity–a module of Toxtree. JRC Rep 43517 1:6
Gini G (2018) QSAR: what else? In: Nicolotti O (ed) Computational toxicology: methods and protocols. Humana Press, New York, NY, pp 79–105
Benfenati E, Golbamaki A, Raitano G, Roncaglioni A, Manganelli S, Lemke F, Norinder U, Lo Piparo E, Honma M, Manganaro A, Gini G (2018) A large comparison of integrated SAR/QSAR models of the Ames test for mutagenicity. SAR QSAR in Environ Res 29(8):591–611. https://doi.org/10.1080/1062936x.2018.1497702
Gini G, Zanoli F, Gamba A, Raitano G, Benfenati E (2019) Could deep learning in neural networks improve the QSAR models? SAR QSAR in Environ Res 30(9):617–642. https://doi.org/10.1080/1062936X.2019.1650827
Gini G, Zanoli F (2020) Machine learning and deep learning methods in ecotoxicological QSAR modeling. In: Roy K (ed) Ecotoxicological QSARs. Springer Nature, Berlin-Heidelberg
Gini G (2020) The QSAR similarity principle in the deep learning era: confirmation or revision? Found Chem 22:383–402. https://doi.org/10.1007/s10698-020-09380-6
Honma M (2020) An assessment of mutagenicity of chemical substances by (quantitative) structure–activity relationship. Genes Environ 42:23. https://doi.org/10.1186/s41021-020-00163-1
Chakravarti SK, Alla SRM (2019) Descriptor free QSAR modeling using deep learning with long short-term memory neural networks. Front Artif Intell. https://doi.org/10.3389/frai.2019.00017
Buckner C, Garson J. (2019) Connectionism. The Stanford Encyclopedia of Philosophy, https://plato.stanford.edu/archives/fall2019/entries/connectionism/
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hung, C., Gini, G. QSAR modeling without descriptors using graph convolutional neural networks: the case of mutagenicity prediction. Mol Divers 25, 1283–1299 (2021). https://doi.org/10.1007/s11030-021-10250-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10250-2