Abstract
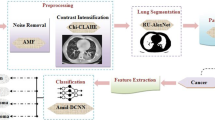
Lung cancer holds a vital spot amidst the causes of cancer deaths among humans. The best way to maximize the survival rate of the patient with cancer is the prior prediction of cancerous cells. But various traditional methods fail to diagnose cancerous cells in their earlier stage. The traditional methods also lead to reduced accuracy. This paper is a mere attempt to demonstrate the methods for diagnosing the disease earlier and also to enhance the accuracy rate. So, at first, the LIDC dataset is considered to be used in this investigation to deal with the higher volume of the scanned image, to attain maximum accuracy. Second, Gaussian noise is excised using Gaussian and Wiener filter. Here pre-processing is carried out to reduce noise in the images. Third, region growing segmentation is used to achieve accurate segmentation of ROI (Region of Interest). In a region growing segmentation, seed points are selected and the adjacent pixels are merged to attain a larger region. After this, features that are extremely significant for a nodule of interest such as area, perimeter, entropy, intensity, or statistical-based features are extracted. From these extracted features, dimensionality is reduced using deep Gaussian mixture model in region-based convolutional neural network [DGMM-RBCNN]. The proposed model is a network with multiple layer latent variables. Here, at every layer, the variables follow Gaussian distributions. Therefore, DGMM forms a cluster of Gaussian distributions to offer a nonlinear model and to describe the image data more flexibly. To eliminate over-parameterized solutions, Gaussian-based dimensionality reduction by designing an overfitting model is used. This is applied at every layer of architecture thereby giving the outcome in deep mixtures of factor analyses. Here, accuracy, sensitivity, specificity, F-measure, ROC curves, and Martins' correlation coefficient are considered as performance metrics. The simulation was carried out in a MATLAB environment, to achieve an accuracy of about 87.79% during the 18thepoch for training and testing the image samples. The false-positive rate could also be determined through this investigation. The anticipated DGMM-RBCNN shows a better and best trade-off than the prevailing systems.

















Similar content being viewed by others
References
Farag AA, Munim HEAE, Graham JH, Farag AA (2013) A novel approach for lung nodules segmentation in chest CT using level sets. IEEE Trans Image Proc 22(12):5202–5213. https://doi.org/10.1109/TIP.2013.2282899
Aryan Mobiny, Supratik Kumar Moulik, Hien Van Nguyen (2018), “Adaptive and Robust Lung Cancer Screening Using Memory-Augmented Recurrent Networks”, https://arxiv.org/pdf/1710.05719.pdf
Teramoto A, Tsukamoto T, Kiriyama Y, Fujita H (2017) Automated classification of lung cancer types from cytological images using deep convolutional neural networks. Hindawi BioMed Res Int. https://doi.org/10.1155/2017/4067832
Baek J, McLachlan G, Flack L (2010) Mixtures of factor analysers with common factor loadings: applications to the clustering and visualization of high-dimensional data. IEEE Trans Pattern Anal Mach Intell 32(7):1298–1309. https://doi.org/10.1109/TPAMI.2009.149
Baudry J-P, Raftery AE, Celeux G, Lo K, Gottardo R (2010) Combining mixture components for clustering. J Comput Graph Stat 19(2):332–353. https://doi.org/10.1198/jcgs.2010.08111
Brooks FJ (2015) Grigsby PW (2015), “Low-order non-spatial effects dominate second order spatial effects in the texture quantifier analysis of 18F-FDG-PET images.” PLoS ONE 10:1–17. https://doi.org/10.1371/journal.pone.0116574
Chen D, Zheng R, Peter D, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu X, He J (2015) Cancer statistics in China. CA- A Cancer J Clinicians 66(2):115–132. https://doi.org/10.3322/caac.21338
Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, Huang CS, Chen CM (2016) “Computer-aided diagnosis with deep learning architecture: applications to breast lesions in us images and pulmonary nodules in CT scans”, https://doi.org/10.1038/srep24454
Elbaz A, Beache GM, Gimelfarb G, Suzuki K, Okada K, Elnakib A, Soliman A, Abdollahi B (2013) Computer-aided diagnosis systems for lung cancer: challenges and methodologies. Int J Biomed Imaging. https://doi.org/10.1155/2013/942353
da Silva GLF, Silva A, de Paiva A, Gattass M (2016) Classification of malignancy of lung nodules in CT images using convolutional neural network. Congresso da Sociedade Brasileira de Computação. https://doi.org/10.5753/sbcas.2016.9894
Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R (2015) Non-small-cell lung cancer. Nat Rev Dis Primers. https://doi.org/10.1038/nrdp.2015.9
Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D (2016) Characterization of PET/CT images using texture analysis: the past, the present… any future? Eur J Nucl Med Mol Imaging. https://doi.org/10.1007/s00259-016-3427-0
He K, Zhang X, Ren S, Sun J (2016), Deep residual learning for image recognition. Conference on Computer Vision and Pattern Recognition (CVPR), pp 770–778, https://doi.org/10.1109/CVPR.2016.90
Hennig C (2010) Methods for merging gaussian mixture components. Adv Data Anal Classif 4(1):3–34. https://doi.org/10.1007/s11634-010-0058-3
Pham HH, Le TT, Tran DQ, Ngo DT, Nguyen HQ (2021) Interpreting chest X-rays via CNNs that exploit hierarchical disease dependencies and uncertainty labels. Neurocomputing 437:186–194. https://doi.org/10.1016/j.neucom.2020.03.127
Wang H, Zhou Z et al (2017) Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res. https://doi.org/10.1186/s13550-017-0260-9
HongQin AiminHao (2018) Multi-view Multi-scale CNNs for lung nodule type classification from CT Images. Pattern Recogn 77:262–275. https://doi.org/10.1016/j.patcog.2017.12.022
Khanfir Kallel S, Almouahed B, Solaiman EB (2018) An iterative possibilistic knowledge diffusion approach for blind medical image segmentation. Pattern Recogn. https://doi.org/10.1016/j.patcog.2018.01.024
Kingsley Kuan et. Al (2013), “Deep Learning for Lung Cancer Detection: Tackling the Kaggle Data Science Bowl 2017 Challenge”, IEEE International Conference on Computer Vision, arxiv:1705.09435
Prabukumar M, Agilandeeswari G (2019) An intelligent lung cancer diagnosis system using cuckoo search optimization and support vector machine classifier. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-017-0655-5
Montanari A, Viroli C (2010) Heteroscedastic factor mixture analysis statistical modelling. Stat Model. https://doi.org/10.1177/1471082X0901000405
Razzak MI, Naz S, Zaib A (2017) Deep learning for medical image processing: overview, challenges and future. Lecture Notes in Computational Vision and Biomechanics. https://doi.org/10.1007/978-3-319-65981-7_12
Muthazhagan R, Rajinigirinath, (2020) An enhanced computer-assisted lung cancer detection method using content based image retrieval and data mining techniques. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02123-7
Chondro P et al (2017) Low order adaptive region growing for lung segmentation on plain chest radiographs. Neurocomputing. https://doi.org/10.1016/j.neucom.2017.09.053
Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ (2015) Machine learning methods for quantitative radiomic biomarkers. Sci Rep. https://doi.org/10.1038/srep13087
Pranjal Sahu, Dantong yu, Mallesham Dasari, Fei Hou, Hong Qin (2019), “A Lightweight Multi-Section CNN for Lung Nodule Classification and Malignancy Estimation”, IEEE Journal of Biomedical and Health Informatics, Vol.23, Issue 3, https://doi.org/10.1109/JBHI.2018.2879834
Song QZ, Zhao L, Luo XingKe (2017) Using deep learning for classification of lung nodules on computed tomography images. Hindawi J Healthcare Eng. https://doi.org/10.1155/2017/8314740
Roth HR, Lu L, Liu J, Yao J, Seff A, Cherry KM et al (2016) Improving computer aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging. https://doi.org/10.1109/TMI.2015.2482920
S Sri Harsha, Anne (2016), “Gaussian Mixture Model and Deep Neural Network based Vehicle Detection and Classification”, International Journal of Advanced Computer Science and Applications, Vol. 7, No. 9, https://doi.org/10.14569/IJACSA.2016.070903
Thakur SK, Singh DP, Choudhary J (2020) Lung cancer identification: a review on detection and classification. Cancer Metastasis Rev 39:989–998. https://doi.org/10.1007/s10555-020-09901-x
Sun W, Zheng B, Qian W (2016) Computer aided lung cancer diagnosis with deep learning algorithms. SPIE Medical Imaging doi 10(1117/12):2216307
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, ErhanD VV, Rabinovich A (2015) Going deeper with convolutions. IEEE Conference on Comput Vision Patt Recogn. https://doi.org/10.1109/CVPR.2015.7298594
Valente IR, Cortez PC, Neto EC, Soares JM, De Albuquerque VH, Tavares JM (2016) Automatic 3D pulmonary nodule detection in CT images: a survey. Comput Methods Programs Biomed. https://doi.org/10.1016/j.cmpb.2015.10.006
Wafaa Alakwaa, Mohammad Nassef, Amr Badr (2017) “Lung Cancer Detection and Classification with 3DConvolutional Neural Network (3D-CNN)”, International Journal of Advanced Computer Science and Applications, https://doi.org/10.14569/IJACSA.2017.080853
Xu-HaoZhi H-B (2018) Saliency driven region-edge-based top down level set evolution reveals the asynchronous focus in image segmentation. Pattern Recogn. https://doi.org/10.1016/j.patcog.2018.03.010
Ying Su, Li D, Chen X (2020) Lung nodule detection based on faster R-CNN framework. Comput Methods Programs Biomed. https://doi.org/10.1109/OJEMB.2020.3023614
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No Conflict of interest has been declared by the author(s).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jena, S.R., George, S.T. & Ponraj, D.N. Lung cancer detection and classification with DGMM-RBCNN technique. Neural Comput & Applic 33, 15601–15617 (2021). https://doi.org/10.1007/s00521-021-06182-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-06182-5