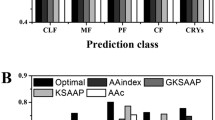
Abstract
Transmembrane proteins play a vital role in cell life activities. There are several techniques to determine transmembrane protein structures and X-ray crystallography is the primary methodology. However, due to the special properties of transmembrane proteins, it is still hard to determine their structures by X-ray crystallography technique. To reduce experimental consumption and improve experimental efficiency, it is of great significance to develop computational methods for predicting the crystallization propensity of transmembrane proteins. In this work, we proposed a sequence-based machine learning method, namely Prediction of TransMembrane protein Crystallization propensity (PTMC), to predict the propensity of transmembrane protein crystallization. First, we obtained several general sequence features and the specific encoded features of relative solvent accessibility and hydrophobicity. Second, feature selection was employed to filter out redundant and irrelevant features, and the optimal feature subset is composed of hydrophobicity, amino acid composition and relative solvent accessibility. Finally, we chose extreme gradient boosting by comparing with other several machine learning methods. Comparative results on the independent test set indicate that PTMC outperforms state-of-the-art sequence-based methods in terms of sensitivity, specificity, accuracy, Matthew's Correlation Coefficient (MCC) and Area Under the receiver operating characteristic Curve (AUC). In comparison with two competitors, Bcrystal and TMCrys, PTMC achieves an improvement by 0.132 and 0.179 for sensitivity, 0.014 and 0.127 for specificity, 0.037 and 0.192 for accuracy, 0.128 and 0.362 for MCC, and 0.027 and 0.125 for AUC, respectively.
Graphic abstract





Similar content being viewed by others
Availability of data and material
The codes and data of PTMC can be obtained from https://github.com/xialab-ahu/PTMC.
Code availability
The codes and data of PTMC can be obtained from https://github.com/xialab-ahu/PTMC.
References
Sweeney MD, Sagare AP, Zlokovic BV (2018) Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol 14(3):133. https://doi.org/10.1038/nrneurol.2017.188
Xu T-H, Yan Y, Kang Y, Jiang Y, Melcher K, Xu HE (2016) Alzheimer’s disease-associated mutations increase amyloid precursor protein resistance to γ-secretase cleavage and the Aβ42/Aβ40 ratio. Cell Discov 2(1):1–14. https://doi.org/10.1038/celldisc.2016.26
Schmit K, Michiels C (2018) TMEM proteins in cancer: a review. Front Pharmacol 9:1345. https://doi.org/10.3389/fphar.2018.01345
Kuhlman B, Bradley P (2019) Advances in protein structure prediction and design. Nat Rev Mol Cell Biol 20(11):681–697. https://doi.org/10.1038/s41580-019-0163-x
Palmer AG, Patel DJ (2002) Kurt Wüthrich and NMR of biological macromolecules. Structure 10(12):1603–1604. https://doi.org/10.1016/s0969-2126(02)00915-2
Nogales E (2015) The development of cryo-EM into a mainstream structural biology technique. Nat Methods 13(1):24. https://doi.org/10.1038/nmeth.3694
Perman B, Anderson S, Schmidt M, Moffat K (2000) New techniques in fast time-resolved structure determination. Cell Mol Biol (Noisy-le-Grand, France) 46(5):895–913
Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J (2000) The Protein Data Bank and the challenge of structural genomics. Nat Struct Mol Biol 7(11s):957. https://doi.org/10.1038/80734
Overton IM, Barton GJ (2006) A normalised scale for structural genomics target ranking: the OB-Score. FEBS Lett 580(16):4005–4009. https://doi.org/10.1016/j.febslet.2006.06.015
Overton IM, Padovani G, Girolami MA, Barton GJ (2008) ParCrys: a Parzen window density estimation approach to protein crystallization propensity prediction. Bioinformatics 24(7):901–907. https://doi.org/10.1093/bioinformatics/btn055
Chen K, Kurgan L, Rahbari M (2007) Prediction of protein crystallization using collocation of amino acid pairs. Biochem Biophys Res Commun 355(3):764–769. https://doi.org/10.1016/j.bbrc.2007.02.040
Kurgan L, Razib AA, Aghakhani S, Dick S, Mizianty M, Jahandideh S (2009) CRYSTALP2: sequence-based protein crystallization propensity prediction. BMC Struct Biol 9(1):50. https://doi.org/10.1186/1472-6807-9-50
Wang H, Feng L, Zhang Z, Webb GI, Lin D, Song J (2016) Crysalis: an integrated server for computational analysis and design of protein crystallization. Sci Rep 6:21383. https://doi.org/10.1038/srep21383
Elbasir A, Moovarkumudalvan B, Kunji K, Kolatkar PR, Mall R, Bensmail H (2019) DeepCrystal: a deep learning framework for sequence-based protein crystallization prediction. Bioinformatics 35(13):2216–2225. https://doi.org/10.1093/bioinformatics/bty953
Mizianty MJ, Kurgan L (2011) Sequence-based prediction of protein crystallization, purification and production propensity. Bioinformatics 27(13):i24–i33. https://doi.org/10.1093/bioinformatics/btr229
Jahandideh S, Mahdavi A (2012) RFCRYS: Sequence-based protein crystallization propensity prediction by means of random forest. J Theor Biol 306:115–119. https://doi.org/10.1016/j.jtbi.2012.04.028
Wang H, Wang M, Tan H, Li Y, Zhang Z, Song J (2014) PredPPCrys: accurate prediction of sequence cloning, protein production, purification and crystallization propensity from protein sequences using multi-step heterogeneous feature fusion and selection. PLoS ONE 9(8):e105902. https://doi.org/10.1371/journal.pone.0105902
Slabinski L, Jaroszewski L, Rychlewski L, Wilson IA, Lesley SA, Godzik A (2007) XtalPred: a web server for prediction of protein crystallizability. Bioinformatics 23(24):3403–3405. https://doi.org/10.1093/bioinformatics/btm477
Jahandideh S, Jaroszewski L, Godzik A (2014) Improving the chances of successful protein structure determination with a random forest classifier. Acta Crystallogr D Biol Crystallogr 70(3):627–635. https://doi.org/10.1107/S1399004713032070
Elbasir A, Mall R, Kunji K, Rawi R, Islam Z, Chuang G-Y, Kolatkar PR, Bensmail H (2019) BCrystal: an interpretable sequence-based protein crystallization predictor. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz762
Varga JK, Tusnády GE (2018) TMCrys: predict propensity of success for transmembrane protein crystallization. Bioinformatics 34(18):3126–3130. https://doi.org/10.1093/bioinformatics/bty342
Chen T, Guestrin C (2016) XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016. ACM, pp 785–794. https://doi.org/10.1145/2939672.2939785
Cover T, Hart P (1967) Nearest neighbor pattern classification. IEEE Trans Inf Theory 13(1):21–27. https://doi.org/10.1109/TIT.1967.1053964
Hearst MA, Dumais ST, Osuna E, Platt J, Scholkopf B (1998) Support vector machines. IEEE Intell Syst Appl 13(4):18–28. https://doi.org/10.1109/5254.708428
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/a:1010933404324
Le Cessie S, Van Houwelingen JC (1992) Ridge estimators in logistic regression. J R Stat Soc Ser C (Appl Stat) 41(1):191–201. https://doi.org/10.2307/2347628
Xia J-F, Zhao X-M, Huang D-S (2010) Predicting protein–protein interactions from protein sequences using meta predictor. Amino Acids 39(5):1595–1599. https://doi.org/10.1007/s00726-010-0588-1
Wang H, Feng L, Webb GI, Kurgan L, Song J, Lin D (2018) Critical evaluation of bioinformatics tools for the prediction of protein crystallization propensity. Brief Bioinform 19(5):838–852. https://doi.org/10.1093/bib/bbx018
Kozma D, Simon I, Tusnady GE (2012) PDBTM: Protein Data Bank of transmembrane proteins after 8 years. Nucleic Acids Res 41(D1):D524–D529. https://doi.org/10.1093/nar/gks1169
Gabanyi MJ, Adams PD, Arnold K, Bordoli L, Carter LG, Flippen-Andersen J, Gifford L, Haas J, Kouranov A, McLaughlin WA (2011) The Structural Biology Knowledgebase: a portal to protein structures, sequences, functions, and methods. J Struct Funct Genom 12(2):45–54. https://doi.org/10.1007/s10969-011-9106-2
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Cai L, Wang L, Fu X, Xia C, Zeng X, Zou Q (2020) ITP-Pred: an interpretable method for predicting, therapeutic peptides with fused features low-dimension representation. Brief Bioinform. https://doi.org/10.1093/bib/bbaa367
Meher PK, Sahu TK, Banchariya A, Rao AR (2017) DIRProt: a computational approach for discriminating insecticide resistant proteins from non-resistant proteins. BMC Bioinform 18(1):1–14. https://doi.org/10.1186/s12859-017-1587-y
Li Q, Zhou W, Wang D, Wang S, Li Q (2020) Prediction of anticancer peptides using a low-dimensional feature model. Front Bioeng Biotechnol 8:892. https://doi.org/10.3389/fbioe.2020.00892
Fu X, Ke L, Cai L, Chen X, Ren X, Gao M (2019) Improved prediction of cell-penetrating peptides via effective orchestrating amino acid composition feature representation. IEEE Access 7:163547–163555. https://doi.org/10.1109/ACCESS.2019.2952738
Chou K-C (2005) Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 21(1):10–19. https://doi.org/10.1093/bioinformatics/bth466
Chou K-C (2009) Pseudo amino acid composition and its applications in bioinformatics. Proteom Syst Biol Curr Proteom 6:262–274. https://doi.org/10.2174/157016409789973707
Cheng J, Randall AZ, Sweredoski MJ, Baldi P (2005) SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res 33(suppl_2):W72–W76. https://doi.org/10.1093/nar/gki396
Hou J, Adhikari B, Cheng J (2018) DeepSF: deep convolutional neural network for mapping protein sequences to folds. Bioinformatics 34(8):1295–1303. https://doi.org/10.1093/bioinformatics/btx780
Rawi R, Mall R, Kunji K, Shen CH, Kwong PD, Chuang GY (2018) PaRSnIP: sequence-based protein solubility prediction using gradient boosting machine. Bioinformatics 34(7):1092–1098. https://doi.org/10.1093/bioinformatics/btx662
Xia C-Q, Pan X, Shen H-B (2020) Protein-ligand binding residue prediction enhancement through hybrid deep heterogeneous learning of sequence and structure data. Bioinformatics 36(10):3018–3027. https://doi.org/10.1093/bioinformatics/btaa110
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157(1):105–132. https://doi.org/10.1016/0022-2836(82)90515-0
Kawashima S, Ogata H, Kanehisa M (1999) AAindex: amino acid index database. Nucleic Acids Res 27(1):368–369. https://doi.org/10.1093/nar/28.1.374
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. https://dl.acm.org/doi/10.5555/1953048.2078195
Cheng N, Li M, Zhao L, Zhang B, Yang Y, Zheng C-H, Xia J (2020) Comparison and integration of computational methods for deleterious synonymous mutation prediction. Brief Bioinform 21(3):970–981. https://doi.org/10.1093/bib/bbz047
Shen Z, Zhang Q, Han K, Huang D-s (2020) A deep learning model for RNA-protein binding preference prediction based on hierarchical LSTM and attention network. IEEE/ACM Trans Comput Biol Bioinf. https://doi.org/10.1109/TCBB.2020.3007544
Li M, Wang Y, Li F, Zhao Y, Liu M, Zhang S, Bin Y, Smith AI, Webb G, Li J (2020) A deep learning-based method for identification of bacteriophage–host interaction. IEEE/ACM Trans Comput Biol Bioinf. https://doi.org/10.1109/TCBB.2020.3017386
Chu Y, Kaushik AC, Wang X, Wang W, Zhang Y, Shan X, Salahub DR, Xiong Y, Wei D-Q (2019) DTI-CDF: a cascade deep forest model towards the prediction of drug-target interactions based on hybrid features. Brief Bioinform. https://doi.org/10.1093/bib/bbz152
Choy BC, Cater RJ, Mancia F, Pryor EE (2021) A 10-year meta-analysis of membrane protein structural biology: detergents, membrane mimetics, and structure determination techniques. Biochim Biophys Acta Biomembr 1863(3):183533. https://doi.org/10.1016/j.bbamem.2020.183533
Acknowledgements
The authors thank the members of our laboratory for their valuable discussions.
Funding
This work was supported by the Anhui Provincial Outstanding Young Talent Support Plan (gxyq2018083), National Natural Science Foundation of China (62072003, 11835014, and U19A2064).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Zhu, Q., Wang, L., Dai, R. et al. Sequence-Based Prediction of Transmembrane Protein Crystallization Propensity. Interdiscip Sci Comput Life Sci 13, 693–702 (2021). https://doi.org/10.1007/s12539-021-00448-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-021-00448-1