Abstract
Alzheimer’s disease is the most common form of dementia, representing 60–70% of dementia cases. The enzyme acetylcholinesterase (AChE) cleaves the ester bonds in acetylcholine and plays an important role in the termination of acetylcholine activity at cholinergic synapses in various regions of the nervous system. The inhibition of acetylcholinesterase is frequently used to treat Alzheimer’s disease. In this study, a merged BindingDB and ChEMBL dataset containing molecules with reported half-maximal inhibitory concentration (IC50) values for AChE (7032 molecules) was used to build machine learning classification models for selecting potential AChE inhibitors from the SistematX dataset (8593 secondary metabolites). A total of seven fivefold models with accuracy above 80% after cross-validation were obtained using three types of molecular descriptors (VolSurf, DRAGON 5.0, and bit-based fingerprints). A total of 521 secondary metabolites (6.1%) were classified as active in this stage. Subsequently, virtual screening was performed, and 25 secondary metabolites were identified as potential inhibitors of AChE. Separately, the crystal structure of AChE in complex with (–)-galantamine was used to perform molecular docking calculations with the entire SistematX dataset. Consensus analysis of both methodologies was performed. Only eight structures achieved combined probability values above 0.5. Finally, two sesquiterpene lactones, structures 15 and 24, were predicted to be able to cross the blood–brain barrier, which was confirmed in the VolSurf+ quantitative model, revealing these two structures as the most promising secondary metabolites for AChE inhibition among the 8593 molecules tested.
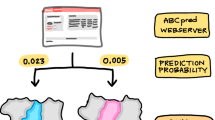
Graphic abstract
A consensus analysis of classification models and molecular docking calculations identified four potential inhibitors of acetylcholinesterase from the SistematX dataset (8593 structures).






Similar content being viewed by others
References
Taylor P, Camp S, Radić Z (2009) Acetylcholinesterase. In: Squire LR (ed) Encyclopedia of neuroscience. Academic Press, Oxford, pp 5–7
Shannon MW, Borron SW, Burn MJ (2007) Chemical weapons. Haddad and Winchester’s clinical management of poisoning and drug overdose (Fourth Edition). W.B. Saunders, Philadelphia, pp 1487–520
Cheung J, Rudolph MJ, Burshteyn F, Cassidy MS, Gary EN, Love J et al (2012) Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J Med Chem 55(22):10282–6. https://doi.org/10.1021/jm300871x
Rotundo RL (2003) Expression and localization of acetylcholinesterase at the neuromuscular junction. J Neurocytol 32(5):743–66. https://doi.org/10.1023/B:NEUR.0000020621.58197.d4
Acetylcholinesterase BD (2007). In: Enna SJ, Bylund DB (eds) xPharm: the comprehensive pharmacology reference. Elsevier, New York, pp 1–8
Akıncıoğlu H, Gülçin İ (2020) Potent acetylcholinesterase inhibitors: potential drugs for Alzheimer’s disease. Mini Rev Med Chem 20(8):703–15. https://doi.org/10.2174/1389557520666200103100521
Mirjana BC, Danijela ZK, Tamara DL-P, Aleksandra MB, Vesna MV (2013) Acetylcholinesterase inhibitors: pharmacology and toxicology. Curr Neuropharmacol 11(3):315–335. https://doi.org/10.2174/1570159X11311030006
Poddar MK, Banerjee S, Chakraborty A, Dutta D (2021) Metabolic disorder in Alzheimer’s disease. Metab Brain Dis. https://doi.org/10.1007/s11011-021-00673-z
Devita M, Masina F, Mapelli D, Anselmi P, Sergi G, Coin A (2021) Acetylcholinesterase inhibitors and cognitive stimulation, combined and alone, in treating individuals with mild Alzheimer’s disease. Aging Clin Exp Res. https://doi.org/10.1007/s40520-021-01837-8
Chekmarev D, Kholodovych V, Kortagere S, Welsh W, Ekins S (2009) Predicting inhibitors of acetylcholinesterase by regression and classification machine learning approaches with combinations of molecular descriptors. Pharm Res 26:2216–24. https://doi.org/10.1007/s11095-009-9937-8
Leuci R, Brunetti L, Poliseno V, Laghezza A, Loiodice F, Tortorella P et al (2021) Natural compounds for the prevention and treatment of cardiovascular and neurodegenerative diseases. Foods 10(1):29. https://doi.org/10.3390/foods10010029
Newman DJ, Cragg GM (2020) Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod 83(3):770–803. https://doi.org/10.1021/acs.jnatprod.9b01285
Calixto JB (2019) The role of natural products in modern drug discovery. Anais da Academia Brasileira de Ciências. https://doi.org/10.1590/0001-3765201920190105
Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K et al (2012) KNApSAcK family databases: integrated metabolite–plant species databases for multifaceted plant research. Plant Cell Physiol 53(2):e1. https://doi.org/10.1093/pcp/pcr165
Banerjee P, Erehman J, Gohlke B-O, Wilhelm T, Preissner R, Dunkel M (2015) Super Natural II—a database of natural products. Nucl Acids Res 43(D1):D935–D9. https://doi.org/10.1093/nar/gku886
Sorokina M, Merseburger P, Rajan K, Yirik MA, Steinbeck C (2021) COCONUT online: Collection of Open Natural Products database. J Cheminformat 13(1):1–13. https://doi.org/10.1186/s13321-020-00478-9
Valli M, Dos Santos RN, Figueira LD, Nakajima CH, Castro-Gamboa I, Andricopulo AD et al (2013) Development of a natural products database from the biodiversity of Brazil. J Nat Prod 76(3):439–44. https://doi.org/10.1021/np3006875
Scotti MT, Herrera-Acevedo C, Oliveira TB, Costa RPO, Santos SYKdO, Rodrigues RP et al (2018) SistematX, an online web-based cheminformatics tool for data management of secondary metabolites. Molecules 23(1):103. https://doi.org/10.3390/molecules23010103
Thomford NE, Senthebane DA, Rowe A, Munro D, Seele P, Maroyi A et al (2018) Natural products for drug discovery in the 21st century: innovations for novel drug discovery. Int J Mol Sci 19(6):1578. https://doi.org/10.3390/ijms19061578
Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK (2007) BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucl Acids Res 35(suppl_1):D198–D201. https://doi.org/10.1093/nar/gkl999
Atanasova M, Yordanov N, Dimitrov I, Berkov S, Doytchinova I (2015) Molecular docking study on galantamine derivatives as cholinesterase inhibitors. Mol Informat 34(6–7):394–403. https://doi.org/10.1002/minf.201400145
Stavrakov G, Philipova I, Zheleva-Dimitrova D, Valkova I, Salamanova E, Konstantinov S et al (2017) Docking-based design and synthesis of galantamine–camphane hybrids as inhibitors of acetylcholinesterase. Chem Biol Drug Design 90(5):709–18. https://doi.org/10.1111/cbdd.12991
Herrera-Acevedo C, Maia MDS, Cavalcanti ÉBVS, Coy-Barrera E, Scotti L, Scotti MT (2011) Selection of antileishmanial sesquiterpene lactones from SistematX database using a combined ligandstructure-based virtual screening approach. Mol Divers. https://doi.org/10.1007/s11030-020-10139-6
Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M et al (2014) QSAR modeling: where have you been? Where are you going to? J Med Chem 57(12):4977–5010. https://doi.org/10.1021/jm4004285
Fourches D, Muratov E, Tropsha A (2015) Curation of chemogenomics data. Nature Chem Biol 11(8):535. https://doi.org/10.1038/nchembio.1881
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–10. https://doi.org/10.1016/S0022-2836(05)80360-2
Cruciani G, Crivori P, Carrupt PA, Testa B (2000) Molecular fields in quantitative structure–permeation relationships: the VolSurf approach. J Mol Struct: THEOCHEM 503(1–2):17–30. https://doi.org/10.1016/S0166-1280(99)00360-7
Cruciani G, Pastor M, Guba W (2000) VolSurf: a new tool for the pharmacokinetic optimization of lead compounds. Eur J Pharm Sci 11:S29–S39. https://doi.org/10.1016/S0928-0987(00)00162-7
Mauri A, Consonni V, Pavan M, Todeschini R (2006) Dragon software: an easy approach to molecular descriptor calculations. Match 56(2):237–48
Todeschini R, Consonni V (2008) Handbook of molecular descriptors. Wiley, New York
Scotti L, Fernandes MB, Muramatsu E, Emereciano VdP, Tavares JF, Silva MSd et al (2011) 13C NMR spectral data and molecular descriptors to predict the antioxidant activity of flavonoids. Braz J Pharm Sci 47(2):241–249. https://doi.org/10.1590/S1984-82502011000200005
Berthold MR, Cebron N, Dill F, Gabriel TR, Kötter T, Meinl T et al (2009) KNIME-the Konstanz information miner: version 2.0 and beyond. AcM SIGKDD Explor Newslett 11(1):26–31. https://doi.org/10.1145/1656274.1656280
Siriseriwan W, Sinapiromsaran K (2016) The effective redistribution for imbalance dataset: relocating safe-level SMOTE with minority outcast handling. Chiang Mai J Sci 43(1):234–46
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143(1):29–36. https://doi.org/10.1148/radiology.143.1.7063747
Matthews BW (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochimica et Biophysica Acta (BBA)-Protein Struct 405(2):442–451. https://doi.org/10.1016/0005-2795(75)90109-9
Scott LJ, Goa KL (2000) Galantamine. Drugs 60(5):1095–1122
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7(1):1–13. https://doi.org/10.1038/srep42717
Sander T, Freyss J, von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55(2):460–73. https://doi.org/10.1021/ci500588j
Crivori P, Cruciani G, Carrupt P-A, Testa B (2000) Predicting blood—brain barrier permeation from three-dimensional molecular structure. J Med Chem 43(11):2204–16. https://doi.org/10.1021/jm990968+
Wong T-T, Yeh P-Y (2019) Reliable accuracy estimates from k-fold cross validation. IEEE Trans Knowl Data Eng 32(8):1586–94. https://doi.org/10.1109/TKDE.2019.2912815
Mukherjee PK, Kumar V, Mal M, Houghton PJ (2007) Acetylcholinesterase inhibitors from plants. Phytomedicine. 14(4):289–300. https://doi.org/10.1016/j.phymed.2007.02.002
Gramatica P, Giani E, Papa E (2007) Statistical external validation and consensus modeling: a QSPR case study for Koc prediction. J Mol Gr Model 25(6):755–66. https://doi.org/10.1016/j.jmgm.2006.06.005
Mues R, Timmermann BN, Ohno N, Mabry TJ (1979) 6-Methoxyflavonoids from Brickellia californica. Phytochemistry 18(8):1379–83. https://doi.org/10.1016/0031-9422(79)83027-7
Bohm BA, Stuessy TF (2001) Flavonoids of the sunflower family (Asteraceae). Springer, Vienna
Baell JB, Holloway GA (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53(7):2719–40. https://doi.org/10.1021/jm901137j
Schmidt TJ (1999) Toxic activities of sesquiterpene lactones: structural and biochemical aspects. Curr Org Chem 3(6):577–608
Seaman FC (1982) Sesquiterpene lactones as taxonomic characters in the Asteraceae. Bot Rev 48(2):121–594. https://doi.org/10.1007/BF02919190
Acevedo CH, Scotti L, Scotti MT (2018) In silico studies designed to select sesquiterpene lactones with potential antichagasic activity from an in-house asteraceae database. ChemMedChem 13(6):634–45
Pérez DJ, Zakai UI, Guo S, Guzei IA, Gómez-Sandoval Z, Razo-Hernández RS et al (2016) Synthesis and biological screening of silicon-containing ibuprofen derivatives: a study of their NF-κβ inhibitory activity, cytotoxicity, and their ability to bind IKKβ. Aust J Chem 69(6):662–71
Funding
We thank the CNPq and Capes for financial Support, Grant Numbers 309648/2019-0 and 431254/2018-4.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Herrera-Acevedo, C., Perdomo-Madrigal, C., Herrera-Acevedo, K. et al. Machine learning models to select potential inhibitors of acetylcholinesterase activity from SistematX: a natural products database. Mol Divers 25, 1553–1568 (2021). https://doi.org/10.1007/s11030-021-10245-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10245-z