Abstract
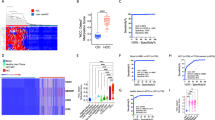
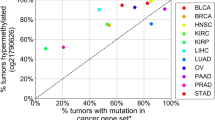
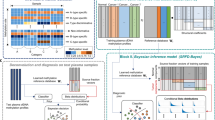
The low abundance of circulating tumour DNA (ctDNA) in plasma samples makes the analysis of ctDNA biomarkers for the detection or monitoring of early-stage cancers challenging. Here we show that deep methylation sequencing aided by a machine-learning classifier of methylation patterns enables the detection of tumour-derived signals at dilution factors as low as 1 in 10,000. For a total of 308 patients with surgery-resectable lung cancer and 261 age- and sex-matched non-cancer control individuals recruited from two hospitals, the assay detected 52–81% of the patients at disease stages IA to III with a specificity of 96% (95% confidence interval (CI) 93–98%). In a subgroup of 115 individuals, the assay identified, at 100% specificity (95% CI 91–100%), nearly twice as many patients with cancer as those identified by ultradeep mutation sequencing analysis. The low amounts of ctDNA permitted by machine-learning-aided deep methylation sequencing could provide advantages in cancer screening and the assessment of treatment efficacy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The microarray data used for identification of differentially methylated sites can be downloaded from the TCGA database at https://gdac.broadinstitute.org/runs/analyses__2016_01_28/data and from the GEO database under the accession code GSE40279. Illumina EPIC TruSeq Methyl data are available at https://basespace.illumina.com/projects/31997005. The raw sequencing data (.fastq files) generated are available from the NCBI Sequence Read Archive (SRA) repository, under the accession code PRJNA534206. The analysed datasets generated during the study are too large to be publicly shared, but they are available for research purposes from the corresponding authors on reasonable request. Any data and materials that can be shared will be released subject to a data-transfer agreement.
Code availability
Codes and scripts developed for this study are available at http://github.com/bnr-ed/mworkflow.
Change history
30 September 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41551-021-00818-6
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Schwarzenbach, H., Hoon, D. S. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011).
Heitzer, E., Perakis, S., Geigl, J. B. & Speicher, M. R. The potential of liquid biopsies for the early detection of cancer. NPJ Precis. Oncol. 1, 36 (2017).
Sun, K. et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl Acad. Sci. USA 112, E5503–E5512 (2015).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
McDonald, B. R. et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 11, eaax7392 (2019).
Libertini, E. et al. Information recovery from low coverage whole-genome bisulfite sequencing. Nat. Commun. 7, 11306 (2016).
Wreczycka, K. et al. Strategies for analyzing bisulfite sequencing data. J. Biotechnol. 261, 105–115 (2017).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer 11, 726–734 (2011).
Dawson, M. A. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science 355, 1147–1152 (2017).
Belinsky, S. A. et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc. Natl Acad. Sci. USA 95, 11891–11896 (1998).
Issa, J. P. CpG island methylator phenotype in cancer. Nat. Rev. Cancer 4, 988–993 (2004).
Guo, S. et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 49, 635–642 (2017).
Hoadley, K. A. et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304.e6 (2018).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005).
Li, W. et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 27, 1243–1257 (2017).
Shen, S. Y. et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018).
Wu, H., Wu, X., Shen, L. & Zhang, Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol. 32, 1231–1240 (2014).
Tanaka, K. & Okamoto, A. Degradation of DNA by bisulfite treatment. Bioorg. Med. Chem. Lett. 17, 1912–1915 (2007).
Bock, C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 13, 705–719 (2012).
Laird, P. W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 11, 191–203 (2010).
Olova, N. et al. Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol. 19, 33 (2018).
Li, J. B. et al. Multiplex padlock targeted sequencing reveals human hypermutable CpG variations. Genome Res. 19, 1606–1615 (2009).
Peng, X. et al. TELP, a sensitive and versatile library construction method for next-generation sequencing. Nucleic Acids Res. 43, e35 (2015).
Raine, A., Manlig, E., Wahlberg, P., Syvänen, A. C. & Nordlund, J. SPlinted Ligation Adapter Tagging (SPLAT), a novel library preparation method for whole genome bisulphite sequencing. Nucleic Acids Res. 45, e36 (2017).
Wu, J., Dai, W., Wu, L. & Wang, J. SALP, a new single-stranded DNA library preparation method especially useful for the high-throughput characterization of chromatin openness states. BMC Genom. 19, 143 (2018).
Hoeijmakers, W. A., Bártfai, R., Françoijs, K. J. & Stunnenberg, H. G. Linear amplification for deep sequencing. Nat. Protoc. 6, 1026–1036 (2011).
Genereux, D. P., Johnson, W. C., Burden, A. F., Stoger, R. & Laird, C. D. Errors in the bisulfite conversion of DNA: modulating inappropriate- and failed-conversion frequencies. Nucleic Acids Res. 36, e150 (2008).
Schirmer, M., D’Amore, R., Ijaz, U. Z., Hall, N. & Quince, C. Illumina error profiles: resolving fine-scale variation in metagenomic sequencing data. BMC Bioinf. 17, 125 (2016).
Vaisvila, R. et al. EM-seq: Detection of DNA methylation at single base resolution from picograms of DNA. Preprint at bioRxiv https://doi.org/10.1101/2019.12.20.884692 (2019).
Haque, I. S. & Elemento, O. Challenges in using ctDNA to achieve early detection of cancer. Preprint at bioRxiv https://doi.org/10.1101/237578 (2017).
Deveson, I. W. et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-00857-z (2021).
Jacobs, B. K., Goetghebeur, E. & Clement, L. Impact of variance components on reliability of absolute quantification using digital PCR. BMC Bioinf. 15, 283 (2014).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Phallen, J. et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9, eaan2415 (2017).
Hajian-Tilaki, K. Sample size estimation in diagnositic test studies of biomedical informatics. J. Biomed. Inf. 48, 193–204 (2014).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
Mazzone, P. J. et al. Evaluating molecular biomarkers for the early detection of lung cancer: when is a biomarker ready for clinical use? An official American Thoracic Society policy statement. Am. J. Respir. Crit. Care Med. 196, e15–e29 (2017).
Hori, S. S. & Gambhir, S. S. Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci. Transl. Med. 3, 109ra116 (2011).
Swanton, C. et al. Prevalence of clonal hematopoiesis of indeterminate potential (CHIP) measured by an ultra-sensitive sequencing assay: exploratory analysis of the Circulating Cancer Genome Atlas (CCGA) study. J. Clin. Oncol. 36, 12003–12003 (2018).
Li, W. et al. CancerDetector: ultrasensitive and non-invasive cancer detection at the resolution of individual reads using cell-free DNA methylation sequencing data. Nucleic Acids Res. 46, e89 (2018).
Snyder, M. W., Kircher, M., Hill, A. J., Daza, R. M. & Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016).
Ebbert, M. T. et al. Evaluating the necessity of PCR duplicate removal from next-generation sequencing data and a comparison of approaches. BMC Bioinf. 17, 239 (2016).
Zhang, T. H., Wu, N. C. & Sun, R. A benchmark study on error-correction by read-pairing and tag-clustering in amplicon-based deep sequencing. BMC Genom. 17, 108 (2016).
Costello, M. et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 41, e67 (2013).
Cristiano, S. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019).
Liu, C. et al. Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers. Nat. Biomed. Eng. 3, 183–193 (2019).
Lennon, A. M. et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 369, eabb9601 (2020).
Xu, R. H. et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 16, 1155–1161 (2017).
Jurmeister, P. et al. Machine learning analysis of DNA methylation profiles distinguishes primary lung squamous cell carcinomas from head and neck metastases. Sci. Transl. Med. 11, eaaw8513 (2019).
Liu, M. C. et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31, 745–759 (2020).
Nassiri, F. et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat. Med. 26, 1044–1047 (2020).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019).
Cancer Stat Facts (Surveillance, Epidemiology and End Results Program (SEER)); https://seer.cancer.gov/statfacts/. Accessed 2020.
Ritchie, M. W. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Faust, G. G. & Hall, I. M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505 (2014).
Li, Y. S. et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Ann. Oncol. 29, 945–952 (2018).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Acknowledgements
We thank K. Kemphues (Cornell University) for critical review of the manuscript. We thank Z. Liang, H. Wu, Z. Jin, F. Tan, S. Chuai, W. Deng, X. Mao, Y. Ma, L. Yang, J. Ye and F. Duan for their assistance with this study. This work was supported, in part, by Beijing Natural Science Foundation (grant number 7182132), the Major Projects of the Beijing Municipal Science and Technology Commission (grant number Z171100002017013), the Capital Special Project for Featured Clinical Application (grant number Z151100004015157), the Peking Union Medical College Hospital Youth Fund (grant numbers PUMCH-2016-2.25, HI626500), and the Peking Union Medical College Special Youth Teacher Project (grant numbers 2014zlgc0717; 2014zlgc0135). We acknowledge research funding, not attached to this study or to any research project or collaboration, from Burning Rock Biotech to the University of California at Berkeley.
Author information
Authors and Affiliations
Contributions
N.L. designed, supervised the clinical study, and provided funding. B.L. designed, supervised the technical study, and wrote the paper. Z.J., P.W., Y. Wang, Y. Wu, Z.C., L.C., Z.B., Hongsheng Liu, L.L., C.H., Y.Q. and Y.C. conducted the clinical study including participant recruitment, sample preparation, clinical information collection and interpretation. C. Wang, T.Z., F.Q., J.S., J. Xu, F.X., H.C., S.F., X.Y, H.H.-Z., J. Xiang and Hao Liu performed the technical development including experiment conduction, computational framework construction and data analysis. C. Wu and X.G. optimized the machine-learning algorithm. H.Z. and S.L designed the clinical study and provided funding. Z.Z. conceived the idea and oversaw the overall direction. All authors discussed the results and contributed to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
T.Z., B.L. and Z.Z. are inventors on a pending patent application held by Burning Rock Biotech related to target deep methylation sequencing (WO2019192489A1, filed in the United States, Canada, Europe, Japan, Sigapore, Australia and Brazil). C. Wang., B.L. and Z.Z. are on a patent application to be submitted by Burning Rock Biotech that covers other aspects of ELSA-seq described in this article.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods and Figs. 1–10.
Supplementary Tables 1–10
Oligonucleotide sequences used in ELSA-seq, methylation-pattern counts, hotspot mutations, clinical metadata and other datasets.
Rights and permissions
About this article
Cite this article
Liang, N., Li, B., Jia, Z. et al. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning. Nat Biomed Eng 5, 586–599 (2021). https://doi.org/10.1038/s41551-021-00746-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00746-5
This article is cited by
-
New regulatory thinking is needed for AI-based personalised drug and cell therapies in precision oncology
npj Precision Oncology (2024)
-
Application of hybrid capsule network model for malaria parasite detection on microscopic blood smear images
Multimedia Tools and Applications (2024)
-
Systematic and benchmarking studies of pipelines for mammal WGBS data in the novel NGS platform
BMC Bioinformatics (2023)
-
Individualized dynamic methylation-based analysis of cell-free DNA in postoperative monitoring of lung cancer
BMC Medicine (2023)
-
Cross-platform comparisons for targeted bisulfite sequencing of MGISEQ-2000 and NovaSeq6000
Clinical Epigenetics (2023)