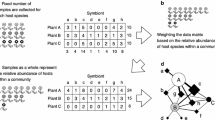
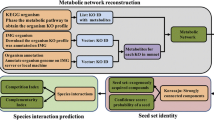
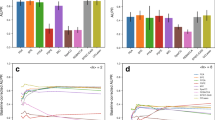
Abstract—
Owing to the high resolving power and efficiency of DNA sequencing, researchers have discovered the extremely high diversity of bacterial, fungal, protist, and microinvertebrate communities in the soil, wood, phyllosphere, and other natural media. Studies on the properties of these communities require powerful tools for analyzing multicomponent systems. One of them is the analysis of ecological networks, which makes it possible to solve a broad range of problems. This review briefly describes the possibilities of network analysis, its concepts, and metrics of network topology.It also indicates limitations related to specific features of DNA sequencing (compositionality and data sparsity) and potential sources of errors in the interpretation of results (relic DNA, artifactual DNA sequences and spurious connections in a network). The focus is on the communities of microorganisms, but the discussed issues are relevant for most other groups of the biota.




Similar content being viewed by others
REFERENCES
Ives, A.R. and Carpenter, S.R., Stability and diversity of ecosystems, Science, 2007, vol. 317, no. 5834, pp. 58–62. https://doi.org/10.1126/science.1133258
Pimm, S.L., The complexity and stability of ecosystems, Nature, 1984, vol. 307, no. 5949, pp. 321–326. https://doi.org/10.1038/307321a0
Tylianakis, J.M., Didham, R.K., Bascompte, J., and Wardle, D.A., Global change and species interactions in terrestrial ecosystems, Ecol. Lett., 2008, vol. 11, no. 12, pp. 1351–1363. https://doi.org/10.1111/j.1461-0248.2008.01250.x
Begon, M., Harper, J.L., and Townsend, C.R., Ecology: Individuals, Populations, and Communities, Oxford: Blackwell, 1986. Translated under the title Ekologiya: Osobi, populyatsii i soobshchestva, Moscow: Mir, 1989.
Layeghifard, M., Hwang, D.M., and Guttman, D.S., Disentangling interactions in the microbiome: A network perspective, Trends Microbiol., 2017, vol. 25, no. 3, pp. 217–228. https://doi.org/10.1016/j.tim.2016.11.008
Nannipieri, P., Ascher, J., Ceccherini, M.T., et al., Microbial diversity and soil functions, Eur. J. Soil Sci., 2003, vol. 54, no. 4, pp. 655–670. https://doi.org/10.1046/j.1351-0754.2003.0556.x
Metzker, M.L., Sequencing technologies – the next generation, Nat. Rev. Genet., 2010, vol. 11, no. 1, pp. 31–46. https://doi.org/10.1038/nrg2626
Bahram, M., Netherway, T., Hildebrand, F., et al., Plant nutrient-acquisition strategies drive topsoil microbiome structure and function, New Phytol., 2020, vol. 227, no. 4, pp. 1189–1199. https://doi.org/10.1111/nph.16598
Davison, J., Moora, M., Öpik, M., et al., Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism, Science, 2015, vol. 349, no. 6251, pp. 970–973. https://doi.org/10.1126/science.aab1161
Tedersoo, L., Bahram, M., Põlme, S., et al., Global diversity and geography of soil fungi, Science, 2014, vol. 346, no. 6213, p. 1078. https://doi.org/10.1126/Science.1256688
Tisthammer, K.H., Cobian, G.M., and Amend, A.S., Global biogeography of marine fungi is shaped by the environment, Fungal Ecol., 2016, vol. 19, pp. 39–46.
Weiss, S., van Treuren, W., Lozupone, C., et al., Correlation detection strategies in microbial data sets vary widely in sensitivity and precision, ISME J., 2016, vol. 10, no. 7, pp. 1669–1681. https://doi.org/10.1038/ismej.2015.235
Faust, K. and Raes, J., Microbial interactions: From networks to models, Nat. Rev. Microbiol., 2012, vol. 10, no. 8, pp. 538–550. https://doi.org/10.1038/nrmicro2832
Newman, M., Networks: An Introduction, Oxford: Oxford Univ. Press, 2010. https://doi.org/10.1093/acprof:oso/9780199206650.001.0001
Jacquiod, S., Puga-Freitas, R., Spor, A., et al., A core microbiota of the plant–earthworm interaction conserved across soils, Soil Biol. Biochem., 2020, vol. 144. https://doi.org/10.1016/j.soilbio.2020.107754
Tipton, L., Muller, C.L., Kurtz, Z.D., et al., Fungi stabilize connectivity in the lung and skin microbial ecosystems, Microbiome, 2018, vol. 6, no. 1, p. 12. https://doi.org/10.1186/s40168-017-0393-0
Jiang, Y.J., Sun, B., Li, H.X., et al., Aggregate-related changes in network patterns of nematodes and ammonia oxidizers in an acidic soil, Soil Biol. Biochem., 2015, vol. 88, pp. 101–109. https://doi.org/10.1016/j.soilbio.2015.05.013
Darcy, J.L., Swift, S.O.I., Cobian, G.M., et al., Fungal communities living within leaves of native Hawaiian dicots are structured by landscape-scale variables as well as by host plants, Mol. Ecol., 2020, vol. 29, pp. 3102–3115. https://doi.org/10.1111/mec.15544
Banerjee, S., Thrall, P.H., Bissett, A., et al., Linking microbial co-occurrences to soil ecological processes across a woodland–grassland ecotone, Ecol. Evol., 2018, vol. 8, no. 16, pp. 8217–8230. https://doi.org/10.1002/ece3.4346
Cram, J.A., Xia, L.C., Needham, D.M., et al., Cross-depth analysis of marine bacterial networks suggests downward propagation of temporal changes, ISME J., 2015, vol. 9, no. 12, pp. 2573–2586. https://doi.org/10.1038/ismej.2015.76
Chaffron, S., Rehrauer, H., Pernthaler, J., and von Mering, C., A global network of coexisting microbes from environmental and whole-genome sequence data, Genome Res., 2010, vol. 20, no. 7, pp. 947–959. https://doi.org/10.1101/gr.104521.109
Meyer, J.L., Gunasekera, S.P., Scott, R.M., et al., Microbiome shifts and the inhibition of quorum sensing by black band disease cyanobacteria, ISME J., 2016, vol. 10, no. 5, pp. 1204–1216. https://doi.org/10.1038/ismej.2015.184
Pollet, T., Berdjeb, L., Garnier, C., et al., Prokaryotic community successions and interactions in marine biofilms: The key role of flavobacteriia, FEMS Microbiol. Ecol., 2018, vol. 94, no. 6. https://doi.org/10.1093/femsec/fiy083
Rottjers, L. and Faust, K., From hairballs to hypotheses: Biological insights from microbial networks, FEMS Microbiol. Rev., 2018, vol. 42, no. 6, pp. 761–780. https://doi.org/10.1093/femsre/fuy030
Deng, Y., Jiang, Y.-H., Yang, Y., et al., Molecular ecological network analyses, BMC Bioinformatics, 2012, vol. 13, no. 1, p. 113. https://doi.org/10.1186/1471-2105-13-113
Dattilo, W. and Rico-Gray, V., Ecological Networks in the Tropics: An Integrative Overview of Species Interactions from Some of the Most Species-rich Habitats on Earth, Cham: Springer, 2018. https://doi.org/10.1007/978-3-319-68228-0.
Junker, B.H. and Schreiber, F., Analysis of Biological Networks, Hoboken, NJ: Wiley, 2008.
Kepes, F., Biological Networks, Singapore: World Scientific, 2007.
Woodward, G., Advances in Ecological Research: Ecological Networks, Amsterdam: Elsevier, 2010.
Semenov, M.V., Metabarcoding and metagenomics in soil-ecological research: Achievements, Problems, and Possibilities, Zh. Obshch. Biol., 2019, vol. 80, no. 6, pp. 403–417. https://doi.org/10.1134/S004445961906006X
Salazar, G., Cornejo-Castillo, F.M., Benitez-Barrios, V., et al., Global diversity and biogeography of deep-sea pelagic prokaryotes, ISME J., 2016, vol. 10, no. 3, pp. 596–608. https://doi.org/10.1038/ismej.2015.137
Pawlowski, J., Audic, S., Adl, S., et al., CBOL protist working group: Barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms, PLoS Biol., 2012, vol. 10, no. 11. https://doi.org/10.1371/journal.pbio.1001419
Mikryukov, V.S., Dulya, O.V., and Modorov, M.V., Phylogenetic signature of fungal response to long-term chemical pollution, Soil Biol. Biochem., 2020, vol. 140, Article no. 107644. https://doi.org/10.1016/j.soilbio.2019.107644
Porter, T.M., Morris, D.M., Basiliko, N., et al., Variations in terrestrial arthropod DNA metabarcoding methods recovers robust beta diversity but variable richness and site indicators, Sci. Rep., 2019, vol. 9, Article no. 18218. https://doi.org/10.1038/s41598-019-54532-0
Chernov, T.I., Zhelezova, O.D., Kutovaya, O.V., et al., Comparative analysis of the structure of buried and surface soils by analysis of microbial DNA, Microbiology (Moscow), 2018, vol. 87, no. 6, pp. 833–841. https://doi.org/10.1134/S0026261718060073
Walters, K.E. and Martiny, J.B.H., Alpha-, beta-, and gamma-diversity of bacteria varies across habitats, PLoS One, 2020, vol. 15, no. 9, e0233872. https://doi.org/10.1371/journal.pone.0233872
Schoch, C.L., Seifert, K.A., Huhndorf, S., et al., Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi, Proc. Natl. Acad. Sci. U. S. A., 2012, vol. 109, no. 16, pp. 6241–6246. https://doi.org/10.1073/pnas.1117018109
Pace, N.R., A molecular view of microbial diversity and the biosphere, Science, 1997, vol. 276, no. 5313, pp. 734–740. https://doi.org/10.1126/science.276.5313.734
Liu, M., Clarke, L.J., Baker, S.C., et al., A practical guide to DNA metabarcoding for entomological ecologists, Ecol. Entomol., 2020, vol. 45, no. 3, pp. 373–385. https://doi.org/10.1111/een.12831
Callahan, B.J., McMurdie, P.J., and Holmes, S.P., Exact sequence variants should replace operational taxonomic units in marker-gene data analysis, ISME J., 2017, vol. 11, no. 12, pp. 2639–2643. https://doi.org/10.1038/ismej.2017.119
Nilsson, R.H., Larsson, K.H., Taylor, A.F.S., et al., The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications, Nucleic Acids Res., 2019, vol. 47, no. D1, pp. D259–D264. https://doi.org/10.1093/nar/gky1022
Pruesse, E., Quast, C., Knittel, K., et al., SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB, Nucleic Acids Res., 2007, vol. 35, no. 21, pp. 7188–7196. https://doi.org/10.1093/nar/gkm864
Guillou, L., Bachar, D., Audic, S., et al., The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy, Nucleic Acids Res., 2013, vol. 41, no. D1, pp. 597–604. https://doi.org/10.1093/nar/gks1160
Ratnasingham, S. and Hebert, P.D., BOLD: The barcode of life data system (http://www.barcodinglife.org), Mol. Ecol. Notes, 2007, vol. 7, no. 3, pp. 355–364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
Gloor, G.B., Macklaim, J.M., Pawlowsky-Glahn, V., and Egozcue, J.J., Microbiome datasets are compositional: and this is not optional, Front. Microbiol., 2017, vol. 8, Article no. 2224. https://doi.org/10.3389/fmicb.2017.02224
Morton, J.T., Marotz, C., Washburne, A., et al., Establishing microbial composition measurement standards with reference frames, Nat. Commun., 2019, vol. 10, no. 1, p. 2719. https://doi.org/10.1038/s41467-019-10656-5
Martin-Fernández, J.-A., Hron, K., Templ, M., et al., Bayesian-multiplicative treatment of count zeros in compositional data sets, Stat. Model., 2015, vol. 15, no. 2, pp. 134–158. https://doi.org/10.1177/1471082X14535524
Freeman, L.C., The Development of Social Network Analysis: A Study in the Sociology of Science, Vancouver BC: Empirical Press, 2004.
Fernandes, A.D., Reid, J.N., Macklaim, J.M., et al., Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16s rRNA gene sequencing and selective growth experiments by compositional data analysis, Microbiome, 2014, vol. 2, p. 15. https://doi.org/10.1186/2049-2618-2-15
Yoon, G., Gaynanova, I., and Muller, C.L., Microbial networks in SPRING – Semi-parametric rank-based correlation and partial correlation estimation for quantitative microbiome data, Front. Genet., 2019, vol. 10, p. 516. https://doi.org/10.3389/fgene.2019.00516
Faust, K., Sathirapongsasuti, J.F., Izard, J., et al., Microbial co-occurrence relationships in the human microbiome, PLoS Comput. Biol., 2012, vol. 8, no. 7. https://doi.org/10.1371/journal.pcbi.1002606
Hirano, H. and Takemoto, K., Difficulty in inferring microbial community structure based on co-occurrence network approaches, BMC Bioinformatics, 2019, vol. 20. https://doi.org/10.1186/s12859-019-2915-1
Csardi, G. and Nepusz, T., The igraph software package for complex network research, InterJournal, Complex Systems, 2006, vol. 1695, no. 5, pp. 1–9.
Bastian, M., Heymann, S., and Jacomy, M., Gephi: An open source software for exploring and manipulating networks, in Proc. Third International AAAI Conf. on Weblogs and Social Media, 2009, vol. 8, pp. 361–362. https://doi.org/10.13140/2.1.1341.1520.
Shannon, P., Markiel, A., Ozier, O., et al., Cytoscape: A software environment for integrated models of biomolecular interaction networks, Genome Res., 2003, vol. 13, no. 11, pp. 2498–2504. https://doi.org/10.1101/gr.1239303
Friedman, J. and Alm, E.J., Inferring correlation networks from genomic survey data, PLoS Comput. Biol., 2012, vol. 8, no. 9. https://doi.org/10.1371/journal.pcbi.1002687
Schwager E., Weingart G., Bielski C. CCREPE: Compositionality corrected by permutation and renormalization. https://www.bioconductor.org/packages/devel/ bioc/html/ccrepe.html.
Reshef, D.N., Reshef, Y.A., Finucane, H.K., et al., Detecting novel associations in large datasets, Science, 2011, vol. 334, no. 6062, pp. 1518–1524. https://doi.org/10.1126/science.1205438
Ruan, Q., Dutta, D., Schwalbach, M.S., et al., Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors, Bioinformatics, 2006, vol. 22, no. 20, pp. 2532–2538. https://doi.org/10.1093/bioinformatics/btl417
Fang, H., Huang, C., Zhao, H., and Deng, M., CCLasso: Correlation inference for compositional data through Lasso, Bioinformatics, 2015, vol. 31, no. 19, pp. 3172–3180. https://doi.org/10.1093/bioinformatics/btv349
Quinn, T.P., Richardson, M.F., Lovell, D., and Crowley, T.M., Propr: An R-package for identifying proportionally abundant features using compositional data analysis, Sci. Rep., 2017, vol. 7. https://doi.org/10.1038/s41598-017-16520-0
Ban, Y., An l., and Jiang, H., Investigating microbial co-occurrence patterns based on metagenomic compositional data, Bioinformatics, 2015, vol. 31, no. 20, pp. 3322–3329. https://doi.org/10.1093/bioinformatics/btv364
Faust, K. and Raes, J., CoNet app: Inference of biological association networks using Cytoscape, F1000Research, 2016, vol. 5. https://doi.org/10.12688/f1000research.9050.2
Antoniazzi, R., Dattilo, W., and Rico-Gray, V., A useful guide of main indices and software used for ecological networks studies, in Ecological Networks in the Tropics, Dattilo, W. and Rico-Gray, V., Eds., Cham: Springer, 2018, pp. 185–196. https://doi.org/10.1007/978-3-319-68228-0_13.
Dunne, J.A., Williams, R.J., and Martinez, N.D., Food-web structure and network theory: The role of connectance and size, Proc. Natl. Acad. Sci. U. S. A., 2002, vol. 99, no. 20, pp. 12917–12922. https://doi.org/10.1073/pnas.192407699
Delmas, E., Besson, M., Brice, M.H., et al., Analyzing ecological networks of species interactions, Biol. Rev., 2019, vol. 94, no. 1, pp. 16–36. https://doi.org/10.1111/brv.12433
Lupatini, M., Suleiman, A.K.A., Jacques, R.J.S., et al., Network topology reveals high connectance levels and few key microbial genera within soils, Front. Environ. Sci., 2014, vol. 2. https://doi.org/10.3389/fenvs.2014.00010
Mikhailov, I.S., Zakharova, Y.R., Bukin, Y.S., et al., Co-occurrence networks among bacteria and microbial eukaryotes of Lake Baikal during a spring phytoplankton bloom, Octolasion cyaneum, Microb. Ecol., 2019, vol. 77, no. 1, pp. 96–109. https://doi.org/10.1007/s00248-018-1212-2
MacArthur R. Fluctuations of animal populations and a measure of community stability, Ecology, 1955, vol. 36, no. 3, pp. 533–536. https://doi.org/10.2307/1929601
May, R.M., Will a large complex system be stable?, Nature, 1972, vol. 238, no. 5364, pp. 413–414. https://doi.org/10.1038/238413a0
Landi, P., Minoarivelo, H.O., Brannstrom, A., et al., Complexity and stability of ecological networks: A review of the theory, Popul. Ecol., 2018, vol. 60, no. 4, pp. 319–345. https://doi.org/10.1007/s10144-018-0628-3
Jalili, M., Salehzadeh-Yazdi, A., Asgari, Y., et al., CentiServer: A comprehensive resource, web-based application and R package for centrality analysis, PLoS One, 2015, vol. 10, no. 11, e0143111. https://doi.org/10.1371/journal.pone.0143111
Lau, M.K., Borrett, S.R., Baiser, B., et al., Ecological network metrics: Opportunities for synthesis, Ecosphere, 2017, vol. 8, no. 8, e01900. https://doi.org/10.1002/ecs2.1900
Faust, K., Bauchinger, F., Laroche, B., et al., Signatures of ecological processes in microbial community time series, Microbiome, 2018, vol. 6, Article no. 120. https://doi.org/10.1186/s40168-018-0496-2
Rottjers, L. and Faust, K., Can we predict keystones?, Nat. Rev. Microbiol., 2019, vol. 17, no. 3, pp. 193–193. https://doi.org/10.1038/s41579-018-0132-y
Agler, M.T., Ruhe, J., Kroll, S., et al., Microbial hub taxa link host and abiotic factors to plant microbiome variation, PLoS Biol., 2016, vol. 14, no. 1, pp. 1–31. https://doi.org/10.1371/journal.pbio.1002352
Douglas, G.M., Maffei, V.J., Zaneveld, J.R., et al., PICRUSt2 for prediction of metagenome functions, Nat. Biotechnol., 2020, vol. 38, no. 6, pp. 685–688. https://doi.org/10.1038/s41587-020-0548-6
Newman, M.E.J., Assortative mixing in networks, Phys. Rev. Lett., 2002, vol. 89, no. 20, 208701. https://doi.org/10.1103/PhysRevLett.89.208701
Yang, Z., Algesheimer, R., and Tessone, C.J., A comparative analysis of community detection algorithms on artificial networks, Sci. Rep., 2016, vol. 6. https://doi.org/10.1038/srep30750
Piraveenan, M., Uddin, S., and Chung, K.S.K., Measuring topological robustness of networks under sustained targeted attacks, in 2012 IEEE/ACM Int. Conf. on Advances in Social Networks Analysis and Mining (Asonam), 2012, pp. 38–45. https://doi.org/10.1109/Asonam.2012.17.
Peel, L., Delvenne, J.C., and Lambiotte, R., Multiscale mixing patterns in networks, Proc. Natl. Acad. Sci. U. S. A., 2018, vol. 115, no. 16, pp. 4057–4062. https://doi.org/10.1073/pnas.1713019115
R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2020. https://www.R-project.org/
Milo, R., Shen-Orr, S., Itzkovitz, S., et al., Network motifs: Simple building blocks of complex networks, Science, 2002, vol. 298, no. 5594, pp. 824–827. https://doi.org/10.1126/science.298.5594.824
Giling, D.P., Ebeling, A., Eisenhauer, N., et al., Plant diversity alters the representation of motifs in food webs, Nat. Commun., 2019, vol. 10, no. 1, p. 1226. https://doi.org/10.1038/s41467-019-08856-0
Stouffer, D.B., Camacho, J., Jiang, W., and Amaral, L.A.N., Evidence for the existence of a robust pattern of prey selection in food webs, Proc. R. Soc. Lond. B, 2007, vol. 274, no. 1621, pp. 1931–1940. https://doi.org/10.1098/rspb.2007.0571
Dell’Anno, A. and Danovaro, R., Extracellular DNA plays a key role in deep-sea ecosystem functioning, Science, 2005, vol. 309, no. 5744, pp. 2179–2179. https://doi.org/10.1126/science.1117475
Carini, P., Delgado-Baquerizo, M., Hinckley, E.L.S., et al., Effects of spatial variability and relic DNA removal on the detection of temporal dynamics in soil microbial communities, mBbio, 2020, vol. 11, no. 1, e02776–02719. https://doi.org/10.1128/mBio.02776-19
Lennon, J.T., Muscarella, M.E., Placella, S.A., and Lehmkuhl, B.K., How, when, and where relic DNA affects microbial diversity, mBio, 2018, vol. 9, no. 3. e00637–00618. https://doi.org/10.1128/mBio.00637-18
Wagner, A.O., Malin, C., Knapp, B.A., and Illmer, P., Removal of free extracellular DNA from environmental samples by ethidium monoazide and propidium monoazide, Appl. Environ. Microbiol., 2008, vol. 74, no. 8, pp. 2537–2539. https://doi.org/10.1128/Aem.02288-07
Baldrian, P., Kolařík, M., Štursová, M., et al., Active and total microbial communities in forest soil are largely different and highly stratified during decomposition, ISME J., 2012, vol. 6, no. 2, pp. 248–258. https://doi.org/10.1038/ismej.2011.95
Edgar, R.C., UNCROSS: Filtering of high-frequency cross-talk in 16S amplicon reads, bioRxiv, 2016, 088666. https://doi.org/10.1101/088666
Davis, N.M., Proctor, D.M., Holmes, S.P., et al., Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data, Microbiome, 2018, vol. 6, Article no. 226. https://doi.org/10.1186/s40168-018-0605-2
Lagkouvardos, I., Fischer, S., Kumar, N., and Clavel, T., Rhea: A transparent and modular R pipeline for microbial profiling based on 16s rRNA gene amplicons, PeerJ, 2017, vol. 5, e2836. https://doi.org/10.7717/peerj.2836
Verhoeven, K.J.F., Simonsen, K.L., and McIntyre, L.M., Implementing false discovery rate control: Increasing your power, Oikos, 2005, vol. 108, pp. 643–647. https://doi.org/10.1111/j.0030-1299.2005.13727.x
Blanchet, F.G., Cazelles, K., and Gravel, D., Co-occurrence is not evidence of ecological interactions, Ecol. Lett., 2020, vol. 23, no. 7, pp. 1050–1063. https://doi.org/10.1111/ele.13525
Berry, D. and Widder, S., Deciphering microbial interactions and detecting keystone species with co-occurrence networks, Front. Microbiol., 2014, vol. 5, Article no. 219. https://doi.org/10.3389/fmicb.2014.00219
Connor, N., Barberan, A., and Clauset, A., Using null models to infer microbial co-occurrence networks, PLoS One, 2017, vol. 12, no. 5, e0176751. https://doi.org/10.1371/journal.pone.0176751
ACKNOWLEDGMENTS
The authors are grateful to M.V. Modorov, O.E. Likho-deevskaya, I.A. Shadrin, and E.A. Belsky for their valuable comments on the manuscript.
Funding
The collection of the material, network construction, bioinformatics analysis, and statistical data processing were supported by the Russian Foundation for Basic Research, project nos. 18-29-05042 and 19-04-00921; the manuscript was prepared for publication under state contract with the Institute of Plant and Animal Ecology, Ural Branch, Russian Academy of Sciences. Bioinformatic analysis was performed using the Uran supercomputer at the Institute of Mathematics and Mechanics, Ural Branch, Russian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest.
Additional information
Translated by N. Gorgolyuk
Rights and permissions
About this article
Cite this article
Mikryukov, V.S., Dulya, O.V., Likhodeevskii, G.A. et al. Analysis of Ecological Networks in Multicomponent Communities of Microorganisms: Possibilities, Limitations, and Potential Errors. Russ J Ecol 52, 188–200 (2021). https://doi.org/10.1134/S1067413621030085
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1067413621030085