Abstract
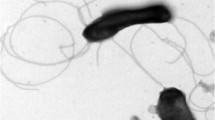
A Gram-negative and rod-shape bacterium designated REN9T, was isolated from pit mud of Baijiu in Sichuan, China. The 16S rRNA sequence of strain REN9T had a high similarity to Pseudoxanthomonas indica P15T (99.21%), P. mexicana AMX 26BT (97.74%) and P. japonensis 12-3T (97.43%). Phylogenetic analysis showed that REN9T belongs to the genus Pseudoxanthomonas and formed distinct cluster. The ANI and DDH values between strains REN9T and P15T were 80.94% and 24%, respectively. Strain REN9T grew optimally at 37 °C, pH 7.0 and 2% NaCl. PE (phosphatidylethanolamine), PG (phosphatidylglycerol) and DPG (diphosphatidylglycerol) were the major polar lipids of REN9T. Ubiquinone 8 (Q-8) was the predominant quinone, and Iso-C15:0 (64.32%), anteiso-C15:0 (12.04%) and Iso-C14:0 (4.56%) were the majority fatty acids. The DNA G + C content of strain REN9T was 67.3 mol%. Genomic analysis showed that strain REN9T had two secondary metabolite biosynthesis gene clusters. Moreover, strain REN9T had 43 glycoside hydrolases, 41 glycosyl transferases and 41 carbohydrate esterases. Based on the polyphasic taxonomic analysis, strain REN9T is recommended as a novel species within the genus Pseudoxanthomonas, for which the name Pseudoxanthomonas beigongshangi is proposed. The type stain is REN9T (= JCM 33961T = GDMCC 1.2210T).




Similar content being viewed by others
Data availability
The genome and 16S rRNA gene of REN9T was submitted to GenBank and got the accession number of JADHDV000000000 and MW187782, respectively.
References
Auch AF, Klenk H, Goeker M (2010) Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2:142–148
Chen MY, Tsay SS, Chen KY et al (2002) Pseudoxanthomonas taiwanensis sp. nov., a novel thermophilic, N2O-producing species isolated from hot springs. Int J Syst Evol Microbiol 52:2155–2161
Finkmann W, Altendorf K, Stackebrandt E, Lipski A (2000) Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int J Syst Evol Microbiol 50(1):273
Fluharty DM (1967) The KOH method for detecting the gram-staining potential of bacteria without staining. Proc Annu Meet US Anim Health Assoc 71:473–479
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Harada RM, Campbell S, Li QX (2006) Pseudoxanthomonas kalamensis sp. nov., a novel gammaproteobacterium isolated from Johnston Atoll, North Pacific Ocean. Int J Syst Evol Microbiol 56:1103–1107
Hu HY, Lim BR, Goto N et al (2001) Analytical precision and repeatability of respiratory quinones for quantitative study of microbial community structure in environmental samples. J Microbiol Methods 47:17–24
Kittiwongwattana C, Thawai C (2016) Pseudoxanthomonas helianthi sp. nov., isolated from roots of Jerusalem artichoke (Helianthus tuberosus). Int J Syst Evol Microbiol 66:5034–5038
Kumari K, Sharma P, Tyagi K et al (2011) Pseudoxanthomonas indica sp. nov., isolated from a hexachlorocyclohexane dumpsite. Int J Syst Evol Microbiol 61:2107–2111
Lee DS, Ryu SH, Hwang HW et al (2008) Pseudoxanthomonas sacheonensis sp. nov., isolated from BTEX-contaminated soil in Korea, transfer of Stenotrophomonas dokdonensis Yoon et al. 2006 to the genus Pseudoxanthomonas as Pseudoxanthomonas dokdonensis comb. nov and emended description of the genus Pseudoxanthomonas. Int J Syst Evol Microbiol 58:2235–2240
Lee JK, Oh JS, Cho WD et al (2017) Pseudoxanthomonas putridarboris sp. nov. isolated from rotten tree. Int J Syst Evol Microbiol 67:1807–1812
Li R, Li Y, Kristiansen K et al (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Li R, Zhu H, Ruan J et al (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Lin J, Yang G, Tang J et al (2019) Pseudoxanthomonas composti sp. nov., isolated from compost. Antonie Van Leeuwenhoek 112:1213–1219
Mohapatra B, Sar P, Kazy SK, Maiti MK, Satyanarayana T (2018) Taxonomy and physiology of Pseudoxanthomonas arseniciresistens sp. Nov., an arsenate and nitrate-reducing novel gammaproteobacterium from arsenic contaminated groundwater, India. PLoS ONE 13:e0193718
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark, DE, USA
Thierry S, Macarie H, Iizuka T (2004) Pseudoxanthomonas mexicana sp. nov. and Pseudoxanthomonas japonensis sp. nov., isolated from diverse environments, and emended descriptions of the genus Pseudoxanthomonas Finkmann et al. 2000 and of its type species. Int J Syst Evol Microbiol 54:2245–2255
Wang GL, Bi M, Liang B et al (2011) Pseudoxanthomonas jiangsuensis sp. nov., a DDT-degrading bacterium isolated from a long-term DDT-polluted soil. Curr Microbiol 62:1760–1766
Weon HY, Kim BY, Kim JS et al (2006) Pseudoxanthomonas suwonensis sp. nov., isolated from cotton waste composts. Int J Syst Evol Microbiol 56:659–662
Xu JL, Sun LP, Sun ZB et al (2019) Pontibacter beigongshangensis sp. nov., isolated from the mash of wine. Curr Microbiol 76:1525–1530
Yoon S, Ha S, Kwon S et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang L, Wei L, Zhu L et al (2014) Pseudoxanthomonas gei sp. nov., a novel endophytic bacterium isolated from the stem of Geum aleppicum. Antonie Van Leeuwenhoek 105:653–661
Acknowledgements
This work supported by The National Key Research and Development Program of China (2018YFC1603606), Earmarked Fund for Hebei Edible Fungi Innovation Team of Modern Agro-industry Technology Research System (HBCT2018050205), and Science and Technology general project of Beijing municipal commission (KM202110011002). We are grateful to Zou Xiaohui, associate researcher at Nationl Institute For Viral Disease Control And Prevention, China CDC with preparing transmission electron microscope.
Funding
This work supported by The National Key Research and Development Program of China (2018YFC1603606), Earmarked Fund for Hebei Edible Fungi Innovation Team of Modern Agro-industry Technology Research System (HBCT2018050205), and Science and Technology general project of Beijing municipal commission (KM202110011002).
Author information
Authors and Affiliations
Contributions
QR designed the experiments. ZBS, FD, LYG and HYG conducted the experiment. JLX and YY contributed to the data analysis. ZBS drafted the manuscript. QR and JLX revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Informed consent
All authors have read and approved the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, Z., Dai, F., Yan, Y. et al. Pseudoxanthomonas beigongshangi sp. nov., a novel bacteria with predicted nitrite and nitrate reduce ability isolated from pit mud of Baijiu. Antonie van Leeuwenhoek 114, 1307–1314 (2021). https://doi.org/10.1007/s10482-021-01603-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01603-w