Abstract
The degree of spatial similarity between the gaze of participants viewing dynamic stimuli such as videos has been previously measured using metrics which are based on the NSS (Normalized Scanpath Saliency). Methods currently used to calculate this metric rely upon a numerical grid, which can be computationally prohibitive for a variety of otherwise useful applications such as Monte Carlo analyses. In the present work we derive a new analytical calculation method for the same metric that yields equal or more accurate results, but with speeds than can be orders of magnitude faster (depending on parameters). Our analytical method scales well with dimensionality, and could also be of use for other applications. The drawback is that it can become very slow if the number of participants in the study is very large or if the gaze sampling rate is high. We provide performance benchmarks for a Fortran implementation of our method, and make available the source code developed.
Similar content being viewed by others
Introduction
Recent technological advances have made gaze tracking practical in a wide variety of settings, both inside and outside of the lab. One particularly promising application for gaze tracking has been to explore how gaze patterns might reveal on-line processing while participants view dynamic stimuli such as videos. For example, researchers have explored how gaze patters might reveal differences in the knowledge or point of view people use to organize their understanding of videos (Huff et al., 2017; Loschky, Larson, Magliano, & Smith, 2015). Differences in gaze patterns between successful and unsuccessful problem solvers when looking at diagrams (Grant & Spivey, 2003) imply that gaze patterns may also reveal differences in learning from videos. One approach to analyzing gaze patterns is to relate them to the visual contents of videos, but it can be difficult to reliably extract valid scene-based regions of interest, calibration issues in real-world settings may preclude effective gaze-object linkages, and it may be impractical to recode each new video one wishes to test. So, an alternative approach is to directly assess the degree to which interesting cognitive similarities between individuals might be reflected in similar gaze patterns across the common time-series of events depicted in the video.
There are many published methods for assessing the similarity between spatio-temporal trajectories. Some have been developed for trajectory data mining purposes, and are very general in terms of the type of data they can be used on. (For some comprehensive reviews of trajectory data mining techniques see Zheng 2015; Atluri et al. 2018 and Bian et al. 2018). Other methods have been developed specifically to compare scanpaths of different viewers, taking into consideration patterns specific to eye fixations and saccadic movements (Anderson, Anderson, Kingstone, & Bischof, 2015; Dorr, Martinetz, Gegenfurtner, & Barth, 2010). Some of these approaches consider similarity between multiple features like position, duration, sequence, amplitudes, and directions (Anderson et al., 2015). However, not all methods are suitable for the analysis of gaze with dynamic stimuli. According to Anderson et al., (2015) the scanpath comparison method should be chosen based on the hypothesis being tested, and for the case of a video, sensitive measures of similarity may be needed. Many approaches such as Scan Path Entropy (Hooge and Camps, 2013), Hidden Markov models (Coutrot, Hsiao, & Chan, 2018), and the popular ScanMatch algorithm (Cristino, Mathôt, Theeuwes, & Gilchrist, 2010), rely on designated regions of interest (ROIs), which may be difficult to establish, especially for experiments involving videos. Additionally, many methods rely on some form of clustering and employ some hard-cutoffs in the process, which could exhibit instabilities due to small perturbations (Dorr et al., 2010), which is an undesirable characteristic.
Currently, a popular approach to linking similarities in gaze patterns with similarities in cognitive variables is to compute similarity metrics which are based on the well-known Normalized Scanpath Saliency (NSS). In general, NSS assesses the degree of correspondence between eyegaze locations on a plane and a scalar field. In many works the scalar field represents variations in perceptual saliency across locations in a visual scene, and NSS has been frequently used to validate computational models of perceptual saliency by comparison to gaze positions (Peters R.J., Iyer A., Itti L., & Koch C., 2005; Riche et al., 2013; Sikha, Kumar, & Soman, 2018).
More recently the NSS has been adapted by Dorr et al., (2010) and other subsequent works (e.g. Loschky et al. 2015and Huff et al. 2017) for the purpose of calculating similarity of gaze scanpaths over time, within and between groups of participants. The goal is to test the degree to which subgroups of participants who might be expected to think differently about a video actually tend to look at different things. If this occurs, then one would expect that the mean similarity of between-participant gaze patterns within groups would be higher than the similarity of gaze patterns between groups (Dorr et al., 2010; Loschky et al., 2015; Lukavský, 2013; Huff et al., 2017; Borji, Sihite, & Itti, 2013). For example, in one study by Huff et al., (2017), fans of two different soccer teams viewed a video of a match between the teams, and NSS was used to test whether fandom affected gaze patterns by assessing the degree to which each fan’s gaze pattern was more similar to other fans of the same team than to fans of the other team. Since the NSS similarity does not require designating regions of interest it is a suitable candidate method for these types of studies (See Dorr et al., 2010, for a more in depth discussion of the advantages of the NSS).
Several variations of NSS-based similarity measures have been used in previous works, but their calculation procedures are closely related. In general, these measures are obtained by first slicing the video into time windows (usually organized around each frame of video), and then obtaining gaze maps and similarity scores for each of those windows (Dorr et al., 2010). Collective gaze maps are constructed from the gaze of groups of participants by superposing Gaussian functions centered at the points of gaze locations (Wooding, 2002; Lukavský, 2013; Dorr et al., 2010). In the example above by Huff et al., (2017), one gaze map was created representing the scanning patterns of fans of soccer team A and another represented fans of team B. The NSS scores were calculated using a leave-one-out procedure in which Gaussians centered on a given participant’s gaze point for each video frame were sampled from a gaze map representing other members of the same fan group, and from the map representing fans of other other team. If fandom drives gaze then we would expect the same-group convolutions would result in higher participant-to-group similarity than between-group convolutions. We note that our reference to “gaze maps” as opposed to “fixation maps” is intentional because these algorithms usually rely on raw gaze positions, rather than the fixations, in order to avoid relying on fixation detection algorithms that may be unreliable for video (Dorr et al., 2010).
Previous works have employed variations with Gaussians that are 2D (e.g. Loschky et al., 2015; Marat et al., 2009), as well as 3D (Dorr et al., 2010; Lukavský, 2013, e.g.). The 3D version considers time as a third dimension, so similarities include both the gaze position at the present time and a small number points before and after, with those that are temporally further away from the present having a lower weight. The 2D version ignores the temporal coordinate and treats all gazepoints within a time window as if they occur simultaneously. An important difference is that the 3D calculation takes into account the temporal ordering of gaze points, while the 2D does not.
One substantial drawback of the methods used in previous works is their reliance upon very computationally expensive calculations for video-like stimuli. The reason for this is that the current calculations of NSS-based metrics employ numerical grids to represent the gaze maps (see Lukavský, 2013), with floating point values for each image pixel, and this is iterated over the many frames in a movie (and iterated again by creating new leave-one-out maps for each participant). The number of pixels in high definition video can be quite large. For example, full resolution 2k video has grid size of 2,073,600 pixels and UHD 4k video has a grid size of up to 8,847,360 pixels. Additionally, for some modern potential applications such as eyetracking in immersive 360 degree VR environments the needed grid could be too large for the method to be practical.
It may be possible to counteract the computational burden in grid calculations by downsampling the data in space or time (sometimes referred to as binning) (Lukavský, 2013). This could come at the cost of accuracy, so it must be done with caution, and the calculations could still be intensive. Also, some applications, such as screen-captured instructional videos, may require fine discriminations between, for example, words and characters that will take advantage of increasing eye tracking accuracy. Thus, a new approach that overcomes these computational burdens while maintaining accuracy is desirable.
There are a number of important reasons to require computational efficiency for NSS calculations. For example, it is often useful to apply Monte Carlo techniques (such as resampling, bootstrapping, or permutation tests), to assess statistical significance. The need for Monte Carlo techniques is invoked in part by the uncertain between-participant independence characteristic of calculations that compare each participant to highly similar leave-one-out maps and by the need to ensure robustness to outliers. Monte Carlo methods can be very powerful and have become more popular recently, for they avoid many of the independence and distributional assumptions inherent to traditional statistical methods, but they require executing many resampling iterations (Manly, 2006). A Monte Carlo approach can typically require 1000 random shuffles (for example, of participant group membership) each of which requires recalculating the entire NSS. Therefore their usage for grid-based calculations can easily become computationally prohibitive.
Caldara and Miellet (2011) analyze 3D Gaussian fixation maps (considering temporal data), using iMap which is an approach inspired by functional magnetic resonance (fMRI) methods. The statistical maps generated by iMap also encounter difficulties due to the high number of pixels. Such limitations were overcome in their work by using random field theory (RFT).
A later version of the work by (Lao, Miellet, Pernet, Sokhn, & Caldara, 2017), iMap4, introduced a pixel-wise linear mixed model (LMM) that is used on Gaussian-smoothed 2D eye fixation maps (no temporal dependence). Monte Carlo statistical tests were employed there as well, and this required some approximations and additional strategies to reduce the large computational burden. Spatial downsampling and masking of pixels were both used by iMap4 to achieve these efficiencies. Lao et al., (2017) also give some guidelines on downsampling.
Eye tracking data can be augmented to very high dimensions with other variables, beyond the x-y coordinates of the gaze (Burch, 2018; Blascheck, Burch, Raschke, & Weiskopf, 2015). For instance, the gaze data acquired can be stereoscopic, and if also taking vergence into consideration then a z dimension needs to be added (Mathôt, Cristino, Gilchrist, & Theeuwes, 2012). Some examples of related ocular data commonly used to augment the raw gaze are : fixation durations, saccade lengths and orientations, pupil dilations (Blascheck et al., 2015; Burch, 2018; Cristino et al., 2010; Mathôt et al., 2012). Also, Blascheck et al., (2015) and Burch (2018) mention that eyetracking data is also being augmented with other sources such as blood pressure, electroencephalography (EEG), functional magnetic resonance imaging (fMRI), galvanic skin response (GSR), mouse and keyboard interactions, motion tracking, verbal data, amongst others. Due to this complexity, some methods have been devised to consider multiple dimensions when analyzing the scanpath (see for example Foerster and Schneider 2013; Cristino et al. 2010; Math\(\hat {\mathrm {o}}\)t et al. 2012; Dewhurst et al. 2012 and Jarodzka et al. 2010). For the case of the calculation of NSS similarity with a grid, high-dimensional gaze data may induce prohibitive computational costs that may further limit the application of this technique.
The new calculation method devised in the present work uses an exact mathematical solution, considering space as continuous rather than discretized, to calculate gaze similarity. It also preserves spatial accuracy regardless of the resolution of the video. We will refer to this approach as the analytical NSS. In short, it yields results with equal or better accuracy than the grid-based approach, and it is capable of running orders of magnitude faster. In the limit of the resolution becoming infinitely large the grid and analytical method return the same scores. Additionally, the analytical NSS can be easily used for gaze data with very high dimensions, allowing its usage for more applications.
We present two forms of the analytical NSS: one to which we refer to as exact, which is limited by computational numerical accuracy, and an approximate form. Both are identical in almost every aspect, and differ only in the equations used to calculate them. Therefore whenever the analytical NSS method is mentioned it is a reference to both forms, unless otherwise specified.
The goal of the present work is to introduce the new analytical NSS approach to calculate gaze similarities. We first present a detailed overview of typical grid-based calculations used in the literature. We then explain our new algorithm and its mathematical formalism. Afterwards, we discuss the usage of Monte Carlo methods with the analytical framework. In the last section we then compare the speed performance of our Fortran implementations of both grid and analytical algorithms, and demonstrate the analytical method’s capability for Monte Carlo calculations.
The overall algorithm
The overall algorithm to calculate the NSS similarity metrics is the same, regardless of whether the calculations are made with the grid-method or the analytical one. In this section we explain what needs to be calculated rather than how, which is left for the later sections detailing each calculation method.
Multiple variations of the NSS similarity metrics have been employed in various studies, although the differences are small, with the core principles from Dorr et al., (2010) being the same. Instead of trying to cover all variations we try to keep the presented formalism in a generalized form so it can be adapted to meet particular needs. The equations presented here assume a 3D calculation, which is easily reducible to the 2D case. Also, we will assume an arbitrary number of groups and participants. We only consider the gaze data from a single eye for each participant (the binocular case is an application with higher dimensionality, for which the method could be extended).
Definitions and notation
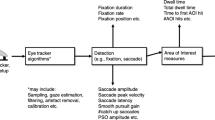
The gaze data are split into time windows, which in general could overlap. Each time window W of duration TW contains all the gaze data between times \(t_{W_{0}}\) and \(t_{W_{f}}=t_{W_{0}}+T_{W}\). Typically the time windows are selected such that they contain data corresponding to one or more video frames. The NSS score is then calculated for each participant with respect to groups of other participants, for each time window separately. A flowchart with the entire NSS calculation for gaze similarity for a single video is shown in Fig. 1.
Let u denote the participant index, \(u \in \{ 1{\dots } n_{u} \} \), where nu is the number of participants, and U the set of all participants. We also define a group of participants g as a subset \(g \subseteq U \).
Groups may be chosen depending on the application, and one typical use would be to account for different experimental conditions. The set of all groups of participants will be denoted as \(\mathcal {G}\).
The participant’s gaze points are denoted as ri,u = (xi,yi,ti), where xi and yi are the horizontal and vertical spatial coordinates (commonly measured in pixels), and ti is the time, which in a 3D calculation is considered as a third dimension. We define Pu as the set of gaze points that correspond to participant u.
We will assume a constant sampling rate for simplicity in the present work (although the algorithms can still be applied if it is not the case). We would then have nt evenly-sampled time points per window for one participant. However, gaze data losses can occur so therefore participant u has Mu ≤ nt gaze points within W. Since there are nu participants there is a total maximum nw = nu × nt gaze points within the window.
For each participant a scalar field \(\mathbb {R}^{3} \!\rightarrow \! \mathbb {R}^{1}\) is constructed consisting of a superposition of 3D Gaussian functions, with each one centered at the participant’s recorded gaze locations in space and time
with
Here \(\nu = \frac {1}{2\sigma ^{2}}\), with σ being the standard deviation of the Gaussian in each dimension. We may assume νx = νy (σx = σy) for simplicity, although it is possible to have arbitrary values.
The gaze map for observer u respect to a reference group of other participants g is then given by the sum of their individual scalar fields:
The raw score η, a scalar, is then obtained by first evaluating the gaze map at the gaze points of participant u that are within the window W, and then averaging those values, as given by
where the sum is done over all valid gaze points of the participant. The raw score can be Z-score normalized by subtracting the mean \(\bar {F}_{u, g } \) and dividing by the standard deviation Std(Fu,g).
A high positive NSS value implies the participant u is looking at a similar location as the others in the reference group, while a low score indicates that the participant is looking at a location where few others are.
The Grid-based calculation
We now describe the outline of a grid calculation (Lukavský, 2013). In this method the scalar field in Eq. 3 is represented using a 3D numerical grid with nx × ny × nt points (or nx × ny for the 2D case), at minimum. In the present work we assume that nx and ny are the number of pixels on the screen in the x and y directions respectively (i.e. no spatial binning is used), unless otherwise specified. Additionally, we assume that in the time dimension there are nt points, (i.e. one gridpoint per gaze sample) which is the minimum needed to avoid temporal binning, unless otherwise indicated. However, in practice one would probably want to have more timepoints for reasons of accuracy, in addition to any required padding during the computation (More details on grid padding can be found in Vetterling and Press, 1992).
An efficient procedure that is typically used to generate a gaze map scalar field is to first create a mask grid with all values initialized to zero, then add 1 to those grid points corresponding to the gaze locations, and afterwards convolve it with another grid containing a single Gaussian function (Tanner & Itti, 2019; Torralba, Castelhano, Oliva, & Henderson, 2006; Lukavský, 2013), which yields as a result a grid with the desired gaze map. Some notes on the implementation of this calculation and its efficiency are given in Appendix B.
Let h be the Gaussian kernel grid and Bg the binary mask formed by the gazepoints of the participants from reference group g, all within a time window. The convolution then yields
One can calculate Fu,g by using Eq. 6 and a convolution routine or a Gaussian filter from existing software libraries. It can also be implemented in an efficient manner using the Fast Fourier Transform (FFT) (Vetterling & Press, 1992). Denoting the FFT and its inverse as \(\mathcal {F}\) and \(\mathcal {F}^{-1}\), and making use of the convolution theorem we therefore have that the scalar field Fu,g in Eq. 3 is then given by
The average value of the grid Fu,g and the standard deviation can be easily calculated, and then by performing a Z-score normalization one obtains the scalar field ηu,g. The NSS score is then obtained by evaluating ηu,g at the gaze point locations of participant u, using Eq. 5.
We summarize the calculation of the NSS score with a grid in Algorithm 1 and Fig. 1.

Algorithm complexity of the grid NSS
Setting the values in the mask to 1 is a very simple operation that must be done once for each Gaussian function in the window, and therefore has complexity \( \mathcal {O}(n_{t}n_{u} ) \). For the convolution process using FFT the computational complexity is \( \mathcal {O}(N\log (N)) \), where N is the total number of gridpoints (Press, Teukolsky, Vetterling, & Flannery, 2007).
The number of gridpoints N will be greatly affected by the dimensionality of the calculation. It will depend of the number of pixels on screen and time points within a window for a 3D calculation (or merely the number of pixels for the 2D case). Unless the number of samples per window or of participants is very large the bottleneck of the entire calculation is likely to be the FFT rather than setting the mask. In that case the computational burden to generate a map for one or many participants is very similar.
For example, for a 2D calculation with a 1600x900 screen (with no spatial downsampling) the number of gridpoints needed is 1.44 million. If we assume a time window with 500 samples and 200 participants we would require setting 100k gridpoints to 1 in the mask. If we consider instead a 3D calculation with the same parameters the number of gridpoints needed is then 720 million, while still having 100k gridpoints to set to 1. In both example cases the bottleneck of the calculation is still the FFT.
The task of setting up the mask might seem like a very repetitive one, and it may be possible to somehow optimize it, although no major improvement is expected unless processing a very large number of participants. Optimization efforts are probably best invested into the usage of strategies to cut down the number of gridpoints (in either time or space) or parallelization.
The analytical calculation
One can make several key observations from the grid algorithm that suggest improvements that serve as the basis for the new method:
-
Many of the operations are sums over all the grid points.
-
All points on the grid need to be computed on all iterations, but many of points are near zero because Gaussians decay rapidly.
-
The leave-one-out procedure is repetitive, and data corresponding to the same participant get introduced or removed together from the calculation.
The calculation can be optimized by assuming space as a continuum and solving some integrals analytically, which avoids the need to sum over every grid point. Also, pre-calculating reduces repetitive storage and retrieval of values. By aggregating these values by participant, the leave-one-out process becomes even more efficient, since fewer operations are needed per iteration.
The overall analytical method of calculation consists of pre-calculating as many values as possible and storing them into arrays (only once per time window), which are then iterated over each participant and reference groups to calculate the NSS scores. Any NSS score involving any arbitrary combination of participants and groups can be obtained using the pre-calculated array values. A large initial investment in computational effort is required, but this makes subsequent iterations within the same window much faster. The analytical method still maintains the coarse structure described by the flowchart in Fig. 1.
Three key quantities are needed to obtain the NSS score, as seen in Eq. 5: the raw score, the average, and the standard deviation. Each of these can be calculated analytically employing a few equations and arrays S, A, θ and M, which are to be pre-calculated once for every time window.
The matrix element \(S[u,u^{\prime }]\) is the total Gaussian similarity between the pair of participants u and \(u^{\prime }\), and it is used to calculate the raw score. Then, A[u] is the contribution of participant u to the average of the gaze map. Also, \(\theta [u,u^{\prime }]\) is the total overlap between the distributions of participant u and \(u^{\prime }\), and is used to calculate the standard deviation of the gaze map. Lastly, M[u] has the number of valid gaze points for participant u within W. A summary of the arrays is given in Table 1, and more detailed descriptions are presented in the upcoming sections, with the full derivations are given in the Appendix. More details of the analytical method of calculation are given in Algorithm 2.
There is no numerical grid representing the gaze map in this method of calculation, so we no longer use nx, ny, and nt. However, the gaze map is still confined within a 3D box defined by the screen size in space and the time window. The sides of this box have lengths Lx, Ly (in spatial units or pixels) and Lt = TW (time units). The volume of the box is then
Raw score
We conveniently define the Gaussian similarity between two gaze points as
which ranges from 0 to 1, with the latter occurring when the two points are identical.
We can also define the Gaussian similarity for a pair of participants, which will be the sum of the similarities of their gaze points. Let ri,u and \(\mathbf {r}_{j,u^{\prime }} \) be the ith and jth gaze points of participants u and \(u^{\prime }\). We then have
where the sum is done over all valid gaze points of those participants within the window.
Using this definition the raw score, Eq. 4, can be rewritten in a form that is more convenient for the analytical method,
Average gaze map - exact form
The contribution of a gaze point ri to the average of the gaze map is
where
and erf is the error function (Abramowitz, 1974).
The total contribution of the participant to the gaze map average is then given by the sum
where the sum goes over all valid gaze points of participant u.
The average of the gaze map of participant u respect to reference group g is then given by
Standard deviation of gaze map - exact form
We now show the equations for the standard deviation of the gaze map, which requires the overlap matrix elements to be calculated. The overlap between two Gaussians centered at gaze points ri and rj is given by
where
and
We can also define an overlap matrix at the participant level, akin to the one shown previously for the Gaussian similarity. This one will has as elements the sum of all the overlaps between all the Gaussians corresponding to a pair of participants. We then have
where the sum is done over all valid gaze points of those participants within the window.
The standard deviation of the gaze map can be obtained from the overlap matrix Eq. 20 and the average field Eq. 15. It is given by
where V is the volume of the box given by Eq. 8.
Approximate forms
A drawback of the analytical method previously presented is that it requires calling the error function multiple times for each matrix element. For a 3D calculation each one requires six error function calls, and four in a 2D case. It is possible to employ an approximation to avoid this if all the gaze points have their center far enough from the boundaries of the box.
Since Gaussians decay very rapidly to zero, if they are far from the screen boundaries the error function approaches its limit value
Using the approximation in Eq. 22 into Eq. 13 gives
Similarly, the overlap integral in Eq. 17 is then simplified to
One can see that this approximation results in a much simpler expression and the error functions are entirely avoided, making the calculation faster, which will be demonstrated in the benchmark section. The algorithm remains exactly the same in every other aspect.
However caution must be taken when using the approximate form, for it could yield an inaccurate result if the Gaussians are too close to the edge of the screen. In that case one possibility is to assume a screen size larger than the actual one to well accommodate all the points, which produces no penalty in the computational burden for the analytical method, although it might affect the meaning of the results. Our code allows the usage of both the exact and approximate forms of the analytical calculation.
Algorithm complexity of the analytical NSS
The computational complexity for the analytical NSS is determined by the number of time points in each window and the number of participants, while being independent of the screen resolution.
The average map array A, from Eq. 14, is one dimensional and has a size nu. One can calculate \(\bar {F}_{u,g }\) by summing only the needed participants within the group. The calculation of the average field requires adding nu terms at the most, and will therefore have a complexity of \( \mathcal {O}(n_{u} )\) operations.
The matrix elements of the similarity and overlap matrices, S and θ, are the ones that will require the largest computation time. Each of these matrices has \({n^{2}_{u}}\) elements. Any NSS score within the time window can be calculated for any group by adding up combinations of these matrix elements, so the process hereby has a complexity \( \mathcal {O}({n^{2}_{u}} )\). However, to pre-calculate all these matrix elements one needs to iterate over all possible pairs of participants and pairs of Gaussians. Since each participant has at most nt gaze points within the window the computational complexity becomes \( \mathcal {O}({{n}_{u}^{2}}{{n}_{t}^{2}} )\). It is therefore possible that the pre-calculation of the matrices will become the bottleneck of the entire process.

Theoretical comparison between grid and analytical methods
A comparison of the computational complexities between the grid and analytical methods is shown in Table 2. The main difference is that in the grid method there is nothing to pre-calculate for each window, for it sets up the binary mask and executes the FFT from scratch every iteration. On the other hand the analytical method makes a large initial investment in computation on each window to obtain the matrix elements, and the gain is that all the subsequent calculations within that window become very fast because they only require a retrieval and summation of these precomputed matrix elements. It becomes quite inexpensive to calculate the similarity score from any arbitrary combination of participants, with any inclusions, exclusions or repetitions needed, such as the leave-one-out procedure. This gain in speed opens the possibility of applying Monte Carlo techniques such as bootstrapping or permutation testing.
Having a quadratic complexity might suggest that the new method could be slow. Nevertheless, the number of participants in many eyetracking studies is usually not very large, < 100, so this method can then become faster than the grid-based calculation by orders of magnitude, as we demonstrate with the benchmarks made with our implementation.
The grid method is strongly affected by dimensionality due to the high number of points required, and extending to higher dimensions can be difficult. On the other hand, the analytical method scales much better with dimensionality and can be easily expanded to additional dimensions, requiring simply another Gaussian integral per dimension to calculate for each matrix element.
The number of time points per window is an important factor affecting speed for both methods. It can vary depending on the sampling rate, which, in the case of research-grade eye trackers, can be in the range 30 − 2000Hz. Since the motions of most interest for dynamic stimuli tend to be fixations and smooth pursuits rather than saccades, sampling rates above 100Hz are often not needed. Video frame rates are typically \(\sim 30 Hz\), and there might be little need to have too many samples per frame, so time downsampling could be a good option to improve performance for both methods, which we later demonstrate.
The exact method also allows one to export the matrices for every time window, which give more information on individual participants, and can be useful for other analyses and data mining algorithms.
Monte Carlo
We now show how the overall algorithm can be generalized to include the application of Monte Carlo methods.
Monte Carlo simulations would only imply extra repetitions of the within-window calculations with some variations in the data employed. Each of these variations is made simply by adding or removing participants to the calculation groups.
For example a permutation test can be done by shuffling the participants into new groups on every Monte Carlo iteration, and repeating the window calculation. A bootstrap calculation would instead require random sampling with replacement.
We can easily generalize the formulation of the calculation for Monte Carlo methods. The flowchart then becomes as shown in Fig. 2.
The theoretical advantage of the analytical NSS here becomes clear because all the intensive computation is pre-calculated once per window, requiring minimal operations to obtain the results in subsequent iterations. However, the random number generation is a process that has not been taken into account in the theoretical formalism, and it can slow down Monte Carlo calculations so the actual performance must be measured in a speed test.
Speed performance benchmarks
We implemented a Fortran code that executes a grid based calculation, as well as the new analytical method in both exact and approximate forms. A big effort was made to make the grid calculations efficient, including the implementation of our own convolution subroutine optimized for this problem. More details on our implementation can be found in Appendix B.
Here we provide a demonstration of the performance of all calculation methods. We used three sets of parameters which are similar to those of some previous studies from Loschky et al., (2015); Dorr et al., (2010) and Marat et al., (2009), in order to make the execution times realistic for future studies in the field. We also created a fourth set of parameters which uses a low sampling rate of 30Hz, which is the approximate video sampling rate. The most relevant parameters for each of these sets are given in Table 3. We denote these parameter sets as A, B, C and D.
The resolution marked with an asterisk (*) in Table 3 has been reduced by a factor of 4 in each dimension respect to the original value. This spatial downsampling (binning) allows the grid calculation to run much faster, and is done merely as a reference for the speed performance, regardless of the accuracy. A comprehensive analysis of the accuracy of the grid approach is beyond the scope of the present work. Nevertheless, a brief test comparing the grid and analytical method scores is presented in Appendix E.
Synthetic gaze data were generated for each run of the benchmark individually, accounting for the number of simulated participants. The purpose of this test is to measure the speed of the methods and ensure correctness of the results, rather than obtaining physiologically accurate ones. See Appendix D for more details on the synthetic data.
Benchmarks were conducted for both 2D and 3D calculations, as well as the case of a Monte Carlo permutation test. In each run, a set of parameters was selected together with a quantity of participants, and then the calculations were done for five time windows. The (simulated) participants were split into two equal groups for each run. All the NSS scores obtained were exported to a file, so file writing times are taken into consideration (these could become more significant for the Monte Carlo calculations).
The reported times only consider the outermost loop depicted in Figs. 1 and 2, which correspond to the calculation of the NSS score the time windows. We excluded the time for global setup operations (which include reading data, array allocations and initializations), since they are only done once per run, regardless of the length of the calculation and not indicative of the algorithm performances. The reported times are the averages per window. All the results presented in the following sections are available in tabular form in the code repository.
2D calculation benchmark
The results for 2D calculations are given in Fig. 3.
Average calculation time per window as a function of the number of participants, for the parameter sets given in Table 3, for all calculation methods
We observe that the grid calculation’s computation time exhibits a linear behavior respect to the number of participants. This is consistent with the complexity shown in Table 2, since the first term is the dominant one due to the high number of gridpoints, and quadratic effects are still not large.
On the other hand the non-linear complexity of the analytical method is evident for all parameter sets. The approximate form can indeed provide a noticeable gain in speed.
The analytical approach is overall faster for parameter sets A and D, and considerably slower for C.
For the set B we have that the performance is not too different if the number of participants is below 50. It is important to notice that this parameter set has spatial downsampling, without which the grid method would be much slower.
For set C we observe that the analytical method is very slow, even for a small number of participants. The reason is because the number of gaze samples per time window is very large, consequence of the high gaze sampling rate, which greatly affects the computational complexity of the analytical approach. The grid method is resistant in this case because it is a 2D calculation.
In set D the computation is fast for all methods due to the small grid and low number of time points per window, which is a consequence of the low gaze sampling rate. This shows the effectiveness of downsampling in both space and time. It is therefore that temporal downsampling of the gaze can be an effective tactic used in conjunction with the analytical method.
3D calculation benchmark
We now show some speed tests doing 3D calculations with the same parameter sets as before, except for C because it has a grid that is very large (Loschky et al., 2015) had similar parameters but only did a 2D calculation. The standard deviation for the temporal dimension, σt, was taken as 26.25ms, and is based on the value used in by Dorr et al., (2010). The results are shown in Fig. 4, and more details on the implementation can be found in Appendix B.
The results for parameter set D for two and three dimensions are shown in Table 4, and a plot of those values corresponding to the analytical calculations is given in Fig. 5 for ease of comparison. It is important to notice that the increase in dimensionality does not cause an immense increase in computation time for the analytical NSS, unlike the grid method. The grid calculation speeds obtained in 3D are about 10 times slower than in 2D, while for the analytical calculations the slowdown is less than a factor of 2. Also, in the 3D case the measured speed of the analytical method is greater than the grid-based by a factor ranging between about 30 to over 500, despite that this grid is low-sampled (else the speed gain would likely be more than 16 times greater).
Comparison 2D vs 3D. Average time per window for the analytical calculations in 2D and 3D, using parameter set D. The values plotted are from Table 4
Monte Carlo benchmark
For testing the Monte Carlo capability of the code we conducted a permutation test for the participant grouping, with 1000 iterations for each window. The calculation for each iteration is the same as in the tests in “2D calculation benchmark”, with the difference being that the groups are randomized on each iteration except for the first one. A flowchart with the Monte Carlo permutation test conducted is shown in Fig. 6.
One of the goals of this benchmark is to measure speed of the code while generating random numbers, which are essential to all Monte Carlo computations. The group randomization is done using a Knuth shuffle, and the random numbers obtained using the well-known Mersenne Twister generator from the MKL library (Intel, 2017). The performance could depend on the choice of generator, and other options exist in MKL, although not tested here. Other Monte Carlo tests are possible (e.g. bootstrap), although the computational requirements should be very similar and therefore we expect these results to generalize in that sense and also be insightful of them.
The results for the Monte Carlo permutation test using the analytical method in exact form, with parameter set D for two-dimensional calculations, are shown in Fig. 7 (It is not done for the grid calculations since they are very slow, and their expected duration is at least 1000 times longer than that of the single window). One can observe in the figure that the calculation with Monte Carlo takes longer, but the execution time does not increase by a factor of 1000 but instead remains low. We also know from the previous results from Fig. 5 that doing the calculation in 3D will still maintain the execution time reasonable.
From the results obtained in Fig. 7 the estimated time for 200 participants for a one minute video at 30 fps is about 12 hours, and even faster if the approximate form is used. A grid calculation would instead take an estimated time of 2000 hours for the same task.
Discussion
The results are consistent with theoretical expectations, and yield efficiencies necessary for practical usage as long as the number of participants or time-points per window is not too large. The purpose of the present work is by no means to displace the grid method, which is clearly superior in some cases, but to instead establish a new option which has other benefits. Our results show that if the number of participants or timepoints per window is very high the analytical method is unfeasible, unless some other optimization or simplification is employed.
The agreement between the scores obtained by the analytical and grid methods is very good. This can be seen in Appendix E, serving as evidence of the correctness of the presented analytical method and the proper functioning of the implemented code.
The results in Table 4 and Fig. 5 demonstrate that the analytical method is very scalable to higher dimensions, and potentially useful for other applications, unlike the grid method. If a higher-dimension grid were to be used then the computation would drastically slow down. Suppose, for example, that a new dimension with 100 points were to be added. It is expected to slow down the calculation by a factor of more than 100. If, instead, one adds in two dimensions with that same number of points then the calculations would be over 10000 times slower, and also requiring more memory, hence rapidly becoming unfeasible. Adding another dimension to the analytical calculations is likely to produce a much smaller slowdown because every added dimension requires two additional error function calls for each matrix element, but the latter do not increase in their number regardless of how many dimensions are present.
Our code was written to keep the flow of the analytical calculations as similar as possible for both exact and approximate forms. The error function call in the code is simply replaced with its limit value given by Eq. 22 in the approximate calculation. We therefore attribute the difference in speed between exact and approximate calculations, observed in Table 4 and Fig. 5, to the error function calls.
It might be possible to optimize the performance of code even further for the approximate case. Aside from the error function, the exponential function call is likely to be one of the most time consuming operations. By exploiting the properties of the exponential functions one can change the product of exponential functions into a single exponential function applied to the algebraic sum of the exponents. By doing so, a single exponential call is needed regardless of how many dimensions exist. Such optimization is not currently done in our implementation due to consistency reasons, in order to assess the impact of the error function usage.
The analytical-approximate form may encounter accuracy difficulties at the edges of the screen. By extending the border of the screen by a few standard deviations this problem can be prevented, which is probably acceptable for many applications. If the stimuli of interest is not near the edges, and since the gazes tend to be center biased (Borji et al., 2013), it is likely that no adjustments will be needed. This is because any gazepoints that are very far from the others will not contribute to the Gaussian similarity nor to the overlap matrix, regardless of how close to the border they may be.
The analytical method is substantially slowed where there are a large number of time points per window, and where there are very large numbers of participants. However, this latter issue is greatly lessened if sampling rates are limited. For example, in a typical use case, researchers might want to analyze data from a large number of participants using commodity-level eye trackers. Not only are these systems limited to relatively slow sampling rates (30-90 Hz), but in many applications researchers are interested in only one gaze location per video frame. In such cases, the analytical method could accommodate hundreds of participants using a 33 ms window with 2-3 samples per window. The average time we obtain for 2000 participants with our code in such scenario is about 47 s per window, and beyond that number the time increases very rapidly (the parameters and results for this calculation are available in the code repository).
Other improvement strategies
It may be possible to further improve the analytical method’s efficiency, since Gaussians are highly localized distributions and therefore many of the matrix elements of S and θ might be very close to zero for those pairs of points very far apart, so these matrices could be treated as sparse hereby cutting down the number of operations required.
Strategies such as spatial hashing (Hastings, Mesit, & Guha, 2005) employed in molecular dynamics simulations (Griebel, Knapek, & Zumbusch, 2007) could significantly reduce quadratic complexities. Some of those strategies are known to be highly effective for gases in which particles are relatively far apart, reducing the quadratic complexity to near linear (See for example Varga & Driscoll, 2011; Griebel et al., 2007 and references within). Also, for some applications another possibility is to use masking (Lao et al., 2017), by only considering points within certain regions of the screen. In applications where the gazepoints are not concentrated these approaches could cut down the number of frames where full NSS calculations are needed.
Potential applications with higher dimensionality
The authors have identified some possible examples of high-dimensional applications where the method could be used.
One example could be data acquired while the participant is in motion, such as that in driving environments (either virtual or real). According to (Braunagel, Geisler, Rosenstiel, & Kasneci, 2017), these cases are problematic for geometric methods and also for methods based on attention maps, and methods such as iMap (Caldara & Miellet, 2011) cannot be applied. The reason is that the regions of interest would need to be recalculated, taking into consideration the vehicle or driver’s motion (Braunagel et al., 2017). The analytical NSS could potentially be used in an application like this one, by including the vehicle or driver’s coordinates as additional spatial dimensions. The greatest gaze similarity is then achieved when the participants look at the same area from the same place.
Another example is the case of immersive videos (Löwe, Stengel, Förster, Grogorick, & Magnor, 2017). These have become possible in recent years due to the availability of 360∘ panoramic cameras, which allow participants to change the field of view by moving their head. By doing so they may be experiencing different stimuli, so there is a need to consider the gaze location as well as the viewing direction (Löwe et al., 2017). That additional variable has prompted Löwe et al., (2017) to develop new methods of analysis beyond the typical ones. We propose extending the analytical NSS to include angular variables so it can be applied in this scenario, by adding in an extra dimension. In such case one must account for the periodic nature of the angular variable, which prompts the usage of a circular distribution for those variables. A Gaussian approximation might be sufficient in many cases (See Jammalamadaka and SenGupta, 1999 ch. 2 for more details), making the formalism presented here adequate.
In studies were participants can scroll or zoom the digital contents of the screen at will it may happen that two of them may be looking at the same feature, but they may be exposed to different content. In a study on reading behaviors by Goodwin et al., (2019) such scenario was encountered, and it resembles the challenge encountered by Löwe et al., (2017). The analytical NSS could also handle this scenario by adding more dimensions to account for the screen visibility, as well as other variables related to reading behaviors from the participants.
The NSS similarity method doesn’t make any assumptions that are specific to gaze data and is vector based, so it may be applicable to other trajectory analysis problems too. This method is also much simpler than others and easier to interpret, although also more limited in scope. One important distinction to highlight is that this method was developed for measuring gaze similarity or hypothesis testing across groups or conditions (e.g. Dorr et al., 2010; Huff et al., 2017), while some of the some of the other existing ones are for discovering trajectory patterns. Nevertheless, there are many common elements. For example, the Gaussian similarity has already been used by Lou et al., (2009) in a trajectory data mining algorithm to determine the probability that two trajectory points match. It is also present in the affinity matrices used in some methods that are clustering-based (Brox and Malik, 2010, e.g.Hu et al., 2007). The presented method is suitable for cases where few moving objects are present, with no known information of where the areas of interest might be, and one is interested in obtaining a measure of proximity. Also this method gives each moving object an individual score indicating proximity, which can be used for other analyses.
Conclusions and remarks
We have presented a new method for calculating the NSS for gaze similarity measurements that yields the same results as the existing grid-based one, retaining all of its statistical properties, but with gains in accuracy and potentially large gains in speed. Not only do these improvements support new iterative analyses such as Monte Carlo approaches, but they also allow the extension of NSS analyses into settings requiring analysis of eye data for lengthy high definition videos because it maintains accuracy and speed regardless of screen resolution.
As theoretically expected, our implementation shows huge speed gains for the analytical calculation compared with the grid method in many, but not all, cases. The grid method remains a better choice if the number of participants or time points per window is large. However, strategies exist that can be used to counteract such obstacles for the analytical calculations.
The approximate form of the analytical NSS produced even larger gains in speed, although care must be taken if a large number of gaze points are near screen boundaries. Therefore, as a precaution we proposed either padding each frame or tracking the number of gaze points within certain number of standard deviations of the frame border if the approximation is to be employed.
The new analytical method presented here also shows very good dimensionality scaling, and it is efficient in either 2D or 3D. This can support new applications of the NSS not only for eye movement data, but possibly for more complex higher-dimensional applications, where the grid method is completely unfeasible. We have identified some specific possible examples of possible applications, and many others may exist involving biological signals such as fMRI and EEG.
The analytical framework presented could be used to further investigate other variations of the NSS measure and make use of the support for high-dimensional data and Monte Carlo methods, in tune with modern trends.
References
Abramowitz, M. (1974) Handbook of mathematical functions, with formulas, graphs, and mathematical tables. New York: Dover Publications, Inc.
Anderson, N.C., Anderson, F., Kingstone, A., & Bischof, W.F. (2015). A comparison of scanpath comparison methods. Behavior Research Methods, 47(4), 1377–1392. https://doi.org/10.3758/s13428-014-0550-3
Atluri, G., Karpatne, A., & Kumar, V. (2018). Spatio-temporal data mining: a survey of problems and methods. ACM Computing Surveys 51(4), 1–41. https://doi.org/10.1145/3161602
Bian, J, Tian, D, Tang, Y, & Tao, D (2018). A survey on trajectory clustering analysis. arXiv:1802.06971
Blascheck, T, Burch, M, Raschke, M, & Weiskopf, D (2015). Challenges and perspectives in big eye-movement data visual analytics. In 2015 Big Data Visual Analytics (BDVA), pp 1–8.
Borji, A., Sihite, D.N., & Itti, L. (2013). Quantitative analysis of human-model agreement in visual saliency modeling: a comparative study. IEEE Transactions on Image Processing, 22(1), 55–69. https://doi.org/10.1109/TIP.2012.2210727
Braunagel, C., Geisler, D., Rosenstiel, W., & Kasneci, E. (2017). Online recognition of driver-activity based on visual scanpath classification. IEEE Intelligent Transportation Systems Magazine, 9(4), 23–36.
Brox, T., & Malik, J. (2010). Object segmentation by long term analysis of point trajectories. In K. Daniilidis, P. Maragos, & N. Paragios (Eds.) Computer vision – ECCV, (Vol. 2010 pp. 282–295). Berlin: Springer.
Burch, M (2018). Identifying similar eye movement patterns with t-sne. In Proceedings of the conference on vision, modeling, and visualization, eurographics association, Goslar, DEU, EG VMV ’18, p 111–118, https://doi.org/10.2312/vmv.20181260
Caldara, R., & Miellet, S. (2011). imap: a novel method for statistical fixation mapping of eye movement data. Behavior Research Methods, 43(3), 864–878. https://doi.org/10.3758/s13428-011-0092-x
Coutrot, A., Hsiao, J.H., & Chan, A.B. (2018). Scanpath modeling and classification with hidden markov models. Behavior Research Methods, 50 (1), 362–379. https://doi.org/10.3758/s13428-017-0876-8
Cristino, F., Mathôt, S, Theeuwes, J., & Gilchrist, I.D. (2010). Scanmatch: A novel method for comparing fixation sequences. Behavior Research Methods, 42(3), 692–700. https://doi.org/10.3758/BRM.42.3.692
Dewhurst, R., Nyström, M., Jarodzka, H., Foulsham, T., Johansson, R., & Holmqvist, K. (2012). It depends on how you look at it: Scanpath comparison in multiple dimensions with multimatch, a vector-based approach. Behavior Research Methods, 44(4), 1079–1100. https://doi.org/10.3758/s13428-012-0212-2
Dorr, M, Martinetz, T, Gegenfurtner, KR, & Barth, E (2010). Variability of eye movements when viewing dynamic natural scenes. Journal of Vision, 10(10), 28–28. https://doi.org/10.1167/10.10.28. https://jov.arvojournals.org/arvo/content_public/journal/jov/932797/jov-10-10-28.pdf
Foerster, RM, & Schneider, WX (2013). Functionally sequenced scanpath similarity method (funcsim): Comparing and evaluating scanpath similarity based on a task’s inherent sequence of functional (action) units. Journal of Eye Movement Research 6(5), 1–22. https://doi.org/10.16910/jemr.6.5.4. https://bop.unibe.ch/JEMR/article/view/2368
Frigo, M, & Johnson, SG (1998). FFTW: An adaptive software architecture for the FFT. In Proceedings of the international conference on acoustics, speech, and signal processing, ICASSP, Seattle, Washington. http://www.fftw.org/, (Vol. 3 pp. 1381–1384).
Goodwin, A.P., Cho, S.J., Reynolds, D., Brady, K., & Salas, J. (2019). Digital versus paper reading processes and links to comprehension for middle school students. American Educational Research Journal, p 0002831219890300. https://doi.org/10.3102/0002831219890300
Grant, ER, & Spivey, MJ (2003). Eye movements and problem solving: Guiding attention guides thought. Psychological Science, 14(5), 462–466. https://doi.org/10.1111/1467-9280.02454, pMID: 12930477.
Griebel, M., Knapek, S., & Zumbusch, G. (2007) Numerical simulation in molecular dynamics: Numerics, algorithms, parallelization, applications texts in computational science and engineering. Germany: Springer-Verlag.
Hastings, E.J., Mesit, J., & Guha, R.K. (2005). Optimization of large-scale, real-time simulations by spatial hashing. In Proc. 2005 summer computer simulation conference, society for modeling & simulation international, new jersey, USA, (Vol. 37 pp. 9–17).
Hooge, I, & Camps, G (2013). Scan path entropy and arrow plots: Capturing scanning behavior of multiple observers. Frontiers in Psychology, 4, 996. https://doi.org/10.3389/fpsyg.2013.00996, https://www.frontiersin.org/article/10.3389/fpsyg.2013.00996
Hu, W., Xie, D., Fu, Z., Zeng, W., & Maybank, S. (2007). Semantic-based surveillance video retrieval. IEEE Transactions on Image Processing, 16(4), 1168–1181.
Huff, M, Papenmeier, F, Maurer, AE, Meitz, TGK, Garsoffky, B, & Schwan, S (2017). Fandom biases retrospective judgments not perception. Scientific Reports, 7, 43083. EP –, https://doi.org/10.1038/srep43083, article.
Intel (2017). Intel math kernel library (mkl). https://software.intel.com/en-us/mkl
Jammalamadaka, S.R., & SenGupta, A. (1999). Topics in circular statistics world scientific publishing co pte ltd, Singapore.
Jarodzka, H, Holmqvist, K, & Nyström, M (2010). A vector-based, multidimensional scanpath similarity measure. In Proceedings of the 2010 Symposium on Eye-Tracking Research & Applications, Association for Computing Machinery, New York, NY, USA, ETRA ’10, p 211–218, https://doi.org/10.1145/1743666.1743718 , https://doi-org.proxy.library.vanderbilt.edu/10.1145/1743666.1743718
Lao, J, Miellet, S, Pernet, C, Sokhn, N, & Caldara, R (2017). imap4: An open source toolbox for the statistical fixation mapping of eye movement data with linear mixed modeling. Behavior Research Methods, 49(2), 559–575. https://doi.org/10.3758/s13428-016-0737-x
Loschky, L.C., Larson, A.M., Magliano, J.P., & Smith, T.J. (2015). What would jaws do? the tyranny of film and the relationship between gaze and higher-level narrative film comprehension. PLOS ONE, 10(11), 1–23. https://doi.org/10.1371/journal.pone.0142474
Lou, Y., Zhang, C., Xie, X., Zheng, Y., Wang, W., & Huang, Y. (2009). Map-matching for low-sampling-rate gps trajectories. In Proceedings of 18th ACM SIGSPATIAL conference on advances in geographical information systems. https://doi.org/https://www.microsoft.com/en-us/research/publication/map-matching-for-low-sampling-rate-gps-trajectories/
Löwe, T, Stengel, M., Förster, E.C., Grogorick, S., & Magnor, M. (2017). Gaze visualization for immersive video. In M. Burch, L. Chuang, B. Fisher, A. Schmidt, & D. Weiskopf (Eds.) Eye tracking and visualization (pp. 57–71). Cham: Springer International Publishing.
Lukavský, J. (2013). Eye movements in repeated multiple object tracking. Journal of Vision, 13(7), 9–9. https://doi.org/10.1167/13.7.9. https://jov.arvojournals.org/arvo/content_public/journal/jov/932812/i1534-7362-13-7-9.pdf
Manly, B. (2006) Randomization bootstrap and monte carlo methods in biology, (3rd edn.) Boca Raton, FL: Chapman & Hall texts in statistical science series, Taylor & Francis.
Marat, S., Ho Phuoc, T., Granjon, L., Guyader, N., Pellerin, D., & Guérin-Dugué, A. (2009). Modelling spatio-temporal saliency to predict gaze direction for short videos. International Journal of Computer Vision, 82(3), 231. https://doi.org/10.1007/s11263-009-0215-3
Mathôt, S., Cristino, F., Gilchrist, I.D., & Theeuwes, J. (2012). A simple way to estimate similarity between pairs of eye movement sequences. Journal of Eye Movement Research 5(1), 1–15. https://doi.org/10.16910/jemr.5.1.4. https://bop.unibe.ch/JEMR/article/view/2326
Peters R.J., Iyer A., Itti L., & Koch C. (2005). Components of bottom-up gaze allocation in natural images. https://doi.org/10.1016/j.visres.2005.03.019. http://www.sciencedirect.com/science/article/pii/S0042698905001975, (Vol. 45 pp. 2397–2416).
Press, W.H., Teukolsky, S.A., Vetterling, W.T., & Flannery, B.P. (2007) Numerical recipes 3rd edition: The art of scientific computing, (3rd edn). New York: Cambridge University Press.
Riche, N, Mancas, M, Duvinage, M, Mibulumukini, M, Gosselin, B, & Dutoit, T (2013). Rare2012: A multi-scale rarity-based saliency detection with its comparative statistical analysis. Signal Processing: Image Communication, 28(6), 642–658. https://doi.org/10.1016/j.image.2013.03.009. http://www.sciencedirect.com/science/article/pii/S0923596513000489
Sikha, O, Kumar, SS, & Soman, K (2018). Salient region detection and object segmentation in color images using dynamic mode decomposition. Journal of Computational Science, 25, 351–366. https://doi.org/10.1016/j.jocs.2017.07.007. http://www.sciencedirect.com/science/article/pii/S1877750317308049
Tanner, J, & Itti, L (2019). A top-down saliency model with goal relevance. Journal of Vision, 19(1), 11–11. https://doi.org/10.1167/19.1.11, https://jov.arvojournals.org/arvo/content_public/journal/jov/937737/i1534-7362-19-1-11.pdf
Torralba, A., Castelhano, M.S., Oliva, A., & Henderson, J.M. (2006). Contextual guidance of eye movements and attention in real-world scenes: the role of global features in object search. Psychological Review, 113, 2006.
Varga, K., & Driscoll, J.A. (2011) Computational nanoscience, (1st edn). USA: Cambridge University Press.
Vetterling, W., & Press, W. (1992) Numerical recipes in FORTRAN: the art of scientific computing, (2nd edn). USA: Fortran numerical recipes, Cambridge University Press.
Wooding, D.S. (2002). Eye movements of large populations: Ii. deriving regions of interest, coverage, and similarity using fixation maps. Behavior Research Methods Instruments, & Computers, 34(4), 518–528. https://doi.org/10.3758/BF03195481
Zheng, Y. (2015). Trajectory data mining: an overview. ACM Trans Intell Syst Technol 6(3), 1–41. https://doi.org/10.1145/2743025
Acknowledgements
The authors would like to thank Prof Amanda Goodwin.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Availability of data and material
The Fotran source code, the benchmark results, the configuration files with the parameters used, and others required to execute the code are available in the Github.com repository https://github.com/jasdevelop/NSS_similarity.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This work was supported by NSF grant 1623625.
Appendices
Appendix A: Mathematical details
Let ϕ be a Gaussian function centered at x0:
A.1 Single Gaussian integral
The integral of a single 1D Gaussian function, centered at x0 in the interval (p,q) is
A.2 Overlap integral
The overlap integral between two Gaussian functions is commonly encountered in many applications such as in quantum mechanics (see for example Varga & Driscoll 2011 and references within), and is one of the core elements for the framework shown in the present work. We here derive its solution.
The overlap integral in 1D for the interval (p,q) for two Gaussians with the same width is given by
The exponent can be rearranged as
We can then express it in quadratic form by defining:
From Eq.7.4.32 in (Abramowitz, 1974) we have
Rearranging terms in the exponent in Eq. 31 and substituting a, b and c:
We conveniently define the quantities
Evaluating the indefinite integral in the interval (p,q) we therefore get the sought result:
It can also be written in terms of the integral of a single Gaussian:
which is the form used in our code since it reuses the routine for the single integral.
A.3 Derivations
In the present section some detailed derivations are presented for the average and standard deviation of the gaze maps.
A.3.1 Average field of gaze map
The average value of a continuous function over the interval (p,q) is given by the integral
where L = q − p, which is the length of the interval.
For the 3D Gaussian function we therefore have that the integral is:
where p,q are the boundaries of the box.
The average field for the continuum case is then given by
So the average scalar field formed by N gazepoints is then given by
where Lx,Ly,Lt are the sizes of the calculation box on each dimension.
For convenience, we employ an array with a size equal to the number of participants nu, which has elements equal to the contribution of participant u to the average scalar field:
We can therefore calculate the average field formed by the gaze of a group of participants g, excluding u, as:
A.3.2 Field Standard deviation
The standard deviation of the scalar field F(x) is given by:
The variance of F(x) can be obtained by integration over the volume of the box V = LxLyLt from
which can also be expressed as:
So the variance can be obtained by calculating the integral of the squared scalar field F(x). If the field is composed by a superposition of N Gaussian functions ψ then
where the sum is done over all the Gaussians. The integral in Eq. 42 is a 3D overlap integral (see Eq. 28).
We then define an overlap matrix, making use of the definition in Eq. 28. It will have a shape of N × N with elements
The standard deviation of the map corresponding to group g can then be written as
We now aggregate the overlap matrix elements by participant, for which we remap each single index to a pair of indices as
where ri,u is the ith gaze point of participant u within the time window. Hence,
We then define an overlap matrix at the participant level which has a shape nu × nu. Each new matrix element is the sum of all overlap matrix elements between the Gaussians of u and \(u^{\prime }\) within the time window. Assuming there are nt gaze points in the time window then the participant-level overlap matrix is given by
where the sum is done over all valid gaze points of those participants within the window. Any missing/invalid gaze points can be omitted from the sum; or equivalently one can consider them as positioned infinitely far away and hence their value is zero and do not contribute.
Since θ is the participant-aggregated form of Θ, the sums of all their matrix elements are equal to each other:
The standard deviation for a field for participant u due to a group g, can therefore be obtained using Eq. 49 and substituting into Eq. 45, but summing only over those pairs of participants within the group and excluding u:
Appendix B: Code Implementation notes
For the present work, we implemented both the grid based NSS and analytical NSS (exact and approximate) in Fortran 2008 for the GNU compiler, which is freely available for all major platforms. There are multiple reasons for such choice of programming language: it is well known for its efficiency and extensively used for scientific computing, it allows easy array manipulations and vectorization, it supports the error function natively, and numerical libraries are available. It can also be difficult to develop efficient code in higher level languages such as Python or MATLAB, where any improvement to the algorithm can easily be undermined by overhead times, possibly invalidating the benchmarks.
We made use of the well-known FFTW (Fastest Fourier Transform in the West)(Frigo & Johnson, 1998), interfaced via Intel’s Math Kernel Library MKL (Intel, 2017). This library is freely available for all major operating systems, and it also provides some well-known random number generators for scientific works. Another candidate language for implementing these algorithms is C/C++, which can also make use of FFTW and MKL.
Despite that the grid-based method can feel more intuitive and appear easier implement, it may not necessarily be the case. Careful considerations such as such as zero-padding, shifting and boundary box delimitation are needed to implement the convolution correctly and efficiently. More detailed guidance for programming FFT-based convolutions can be found in Vetterling and Press (1992) and Press et al., (2007) for Fortran and C/C++ respectively. The authors spent considerably more time developing and debugging the grid calculation routines than the analytical ones.
It can be possible to calculate Eq. 7 easily using Gaussian filters or convolution operations from existing software libraries, depending on what language is employed. However, one can reduce the computation time by implementing the convolution operation by oneself, which we did in the present work. One reason is that one can ensure that all the arrays are allocated at the start and avoid repetitive memory allocations during the calculations. Also, the grid with the Fourier Transform of the Gaussian kernel, \(\mathcal {F}(h)\) (see Eq. 7), is has a constant value during the entire video, so it only needs to be created once at the beginning of the video and stored. If so then one only requires two calls to the FFT (one forward and one inverse) instead of three calls for each gaze map. Since the FFT is the most likely bottleneck the improvement in speed can be noticeable.
Another issue that affects performance is memory usage. FFTW has routines that use the same input array as output (in-place operation), which is more efficient than having to copy the array. Once the statistics of the gaze map are calculated there is no need to store it anymore, and there is no need to simultaneously access the binary mask, so these arrays can be shared and therefore one can employ in-place operations.
The FFTW library has different algorithms available. It is certainly possible to make use of the different ones depending on the specific parameters of the calculation in order to gain some speed, but for simplicity and consistency a 3D-complex to 3D-complex FFT was used, which suits all possible scenarios studied in the present work.
The results presented here are normalized respect to the NSS score of a single Gaussian, in a similar manner as done by Dorr et al., (2010). The single Gaussian is placed in the center of the box in our code, for both grid and analytical methods. This calculation is done once and the result stored.
Appendix C: Hardware notes
All the calculations shown here were done using a personal computer with an Intel Core i5 processor, on a Kubuntu 16 Linux distribution running on an Oracle VM Virtual Box. The authors have previously tested the performance of other scientific computing applications and have found that the execution times are comparable to those obtained on a native operating system.
Appendix D: Synthetic data
A simple scenario was crafted to test out the speed of the calculations. The synthetic gaze data were made to have a rectilinear uniform motion across the screen, with individual participants constrained to separate equally-spaced parallel horizontal lanes (reminiscent of an Olympic swimming pool). The gazes move at a speed such that they cross the screen width in five time windows. The number of iterations of the code is exactly the same regardless of the trajectories selected for the synthetic gaze data.
One could expect some possible variability in the number of floating point operations (FLOPS) due to the exponential and error function calls, which could have some dependence on the values calculated. To ensure our benchmarks are not strongly affected by this, the calculations for parameter set D were repeated with random gaze position values. The execution times differed by 3% or less, hence we do not expect the synthetic data to give unrealistic results respect to real data.
Appendix E: Accuracy
In order to compare the NSS scores another calculation was done, using some simulated data, as an accuracy test to demonstrate the agreement between the grid and analytical methods. The NSS scores for the case of two observers, one at the center of the screen and the other separated by a distance d, were calculated with the analytical-exact and grid methods in 2D and 3D. The standard deviation σx, was adjusted such that it corresponds to 1/50 of the screen’s horizontal length. The temporal sampling rate was set to 30 Hz and the screen resolutions of 320x180 and 1280x720 pixels were used. The full simulation parameters are available in the repository.
The results are plotted in Fig. 8, and a table with the results is available in the repository. The distance d has been normalized respect to the standard deviation σx.
NSS scores for various separations, in units of σx, for both the grid and analytical-exact methods, in 2D and 3D (left and right respectively), for low and high spatial resolution (top and bottom). In general the values obtained with both methods are in agreement. Some small discretization artifacts are more evident for low resolution
In general there is very good agreement between the scores, and hence the red and blue markers in the plot are largely overlapping, with some deviations. One can observe some small discretization artifacts, in the manner of the step-like patterns, for the low resolution case. These artifacts are not evident anymore for the high resolution results presented, which also display smaller differences between both methods, as expected.
For the 3D case the scores are overall lower than for 2D as a consequence of the existence of the temporal dimension, which increases the volume of the box, and because the gaze is always moving along the time axis. Also, the grid method is in essence doing a numerical integration, so the inclusion of the temporal axis is therefore expected to introduce more error, which is indeed observed here (note that in the presented calculations only the spatial resolution is different, with the temporal being the same). At short distances the agreement between methods is higher for the 2D case than for 3D, with the difference obtained being about 3% for the latter, which may be reasonable for many purposes. It can be improved by increasing the number of time gridpoints (not done here). Nevertheless, one is typically interested in doing comparisons between scores, hence systematic errors may cancel out and work well in practice.
Note that the results shown in the present section are only meant to be a guideline, for they correspond to this specific scenario and parameters, and could vary for other circumstances. Acceptable error levels are likely to be dependent on the specific application for which the method is used.
The agreement between the calculations serves as evidence of the validity of the analytical method, and that the implemented code is functioning correctly. The calculations for both grid and analytical methods are done by the code using separate modules and functions, with the common part being the input loading portions, which further sustains the validity.
Rights and permissions
About this article
Cite this article
Salas, J.A., Levin, D.T. Efficient calculations of NSS-based gaze similarity for time-dependent stimuli. Behav Res 54, 94–116 (2022). https://doi.org/10.3758/s13428-021-01562-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3758/s13428-021-01562-0