Abstract
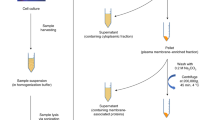
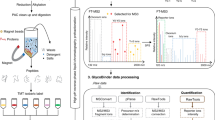
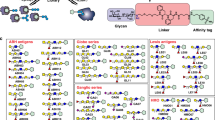
Glycosphingolipids (GSLs) are ubiquitous glycoconjugates present on the cell membrane; they play significant roles in many bioprocesses such as cell adhesion, embryonic development, signal transduction and carcinogenesis. Analyzing such amphiphilic molecules is a major challenge in the field of glycosphingolipidomics. We provide a step-by-step protocol that uses a lectin microarray to analyze GSL glycans from cultured cells. The procedure describes (i) extraction of GSLs from cell pellets, (ii) N-monodeacylation using sphingolipid ceramide N-deacylase digestion to form lyso-GSLs, (iii) fluorescence labeling at the newly exposed amine group, (iv) preparation of a lectin microarray, (v) GSL-glycan analysis by a lectin microarray, (vi) complementary mass spectrometry analysis and (vii) data acquisition and analysis. This method is high-throughput, low cost and easy to conduct, and it provides detailed information about glycan linkages. This protocol takes ~10 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that all the data generated or analyzed during this study are available within the protocol. Source data are provided with this paper.
References
Levery, S. B. Glycosphingolipid structural analysis and glycosphingolipidomics. Methods Enzymol. 405, 300–369 (2005).
D’Angelo, G., Capasso, S., Sticco, L. & Russo, D. Glycosphingolipids: synthesis and functions. FEBS J. 280, 6338–6353 (2013).
Dauner, M., Batroff, E., Bachmann, V., Hauck, C. R. & Wittmann, V. Synthetic glycosphingolipids for live-cell labeling. Bioconjug. Chem. 27, 1624–1637 (2016).
Platt, F. M. Sphingolipid lysosomal storage disorders. Nature 510, 68–75 (2014).
Shayman, J. A. & Larsen, S. D. The development and use of small molecule inhibitors of glycosphingolipid metabolism for lysosomal storage diseases. J. Lipid Res. 55, 1215–1225 (2014).
Ho, M. Y., Yu, A. L. & Yu, J. Glycosphingolipid dynamics in human embryonic stem cell and cancer: their characterization and biomedical implications. Glycoconj. J. 34, 765–777 (2017).
Woeste, M. A. & Wachten, D. The enigmatic role of GBA2 in controlling locomotor function. Front. Mol. Neurosci. 10, 386 (2017).
Zhao, X. et al. Lipidomic profiling links the Fanconi anemia pathway to glycosphingolipid metabolism in head and neck cancer cells. Clin. Cancer Res. 24, 2700–2709 (2018).
Fujiwara, Y., Hama, K. & Yokoyama, K. Mass spectrometry in combination with a chiral column and multichannel-MRM allows comprehensive analysis of glycosphingolipid molecular species from mouse brain. Carbohydr. Res. 490, 107959 (2020).
Ivleva, V. B. et al. Coupling thin-layer chromatography with vibrational cooling matrix-assisted laser desorption/ionization Fourier transform mass spectrometry for the analysis of ganglioside mixtures. Anal. Chem. 76, 6484–6491 (2004).
Kyogashima, M. et al. Rapid demonstration of diversity of sulfatide molecular species from biological materials by MALDI-TOF MS. Glycobiology 16, 719–728 (2006).
Gabius, H. J., Andre, S., Jimenez-Barbero, J., Romero, A. & Solis, D. From lectin structure to functional glycomics: principles of the sugar code. Trends Biochem. Sci. 36, 298–313 (2011).
Du, H. et al. Analysis of glycosphingolipid glycans by lectin microarrays. Anal. Chem. 91, 10663–10671 (2019).
Pilobello, K. T. & Mahal, L. K. Lectin microarrays for glycoprotein analysis. Methods Mol. Biol. 385, 193–203 (2007).
Yu, H., Shu, J. & Li, Z. Lectin microarrays for glycoproteomics: an overview of their use and potential. Expert Rev. Proteomics 17, 27–39 (2020).
Hsu, K. L. & Mahal, L. K. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat. Protoc. 1, 543–549 (2006).
Kuno, A. et al. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nat. Methods 2, 851–856 (2005).
Charles, P. T. et al. Fabrication and characterization of 3D hydrogel microarrays to measure antigenicity and antibody functionality for biosensor applications. Biosens. Bioelectron. 20, 753–764 (2004).
Hirabayashi, J., Yamada, M., Kuno, A. & Tateno, H. Lectin microarrays: concept, principle and applications. Chem. Soc. Rev. 42, 4443–4458 (2013).
Angeloni, S. et al. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology 15, 31–41 (2005).
Pilobello, K. T., Krishnamoorthy, L., Slawek, D. & Mahal, L. K. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem 6, 985–989 (2005).
Hsu, K. L., Pilobello, K. T. & Mahal, L. K. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat. Chem. Biol. 2, 153–157 (2006).
Krishnamoorthy, L., Bess, J. W. Jr., Preston, A. B., Nagashima, K. & Mahal, L. K. HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nat. Chem. Biol. 5, 244–250 (2009).
Hiono, T. et al. Lectin microarray analyses reveal host cell-specific glycan profiles of the hemagglutinins of influenza A viruses. Virology 527, 132–140 (2019).
Zheng, T., Peelen, D. & Smith, L. M. Lectin arrays for profiling cell surface carbohydrate expression. J. Am. Chem. Soc. 127, 9982–9983 (2005).
Pilobello, K. T., Slawek, D. E. & Mahal, L. K. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc. Natl Acad. Sci. USA 104, 11534–11539 (2007).
Tateno, H. et al. A novel strategy for mammalian cell surface glycome profiling using lectin microarray. Glycobiology 17, 1138–1146 (2007).
Hsu, K. L., Pilobello, K., Krishnamoorthy, L. & Mahal, L. K. Ratiometric lectin microarray analysis of the mammalian cell surface glycome. Methods Mol. Biol. 671, 117–131 (2011).
Bird-Lieberman, E. L. et al. Molecular imaging using fluorescent lectins permits rapid endoscopic identification of dysplasia in Barrett’s esophagus. Nat. Med. 18, 315–321 (2012).
Matsuda, A. et al. Development of an all-in-one technology for glycan profiling targeting formalin-embedded tissue sections. Biochem. Biophys. Res. Commun. 370, 259–263 (2008).
Tozawa-Ono, A. et al. Glycan profiling using formalin-fixed, paraffin-embedded tissues: Hippeastrum hybrid lectin is a sensitive biomarker for squamous cell carcinoma of the uterine cervix. J. Obstet. Gynaecol. Res. 43, 1326–1334 (2017).
Zou, X. et al. A standardized method for lectin microarray-based tissue glycome mapping. Sci. Rep. 7, 43560 (2017).
Hu, S. & Wong, D. T. Lectin microarray. Proteomics Clin. Appl. 3, 148–154 (2009).
Narimatsu, H. et al. A strategy for discovery of cancer glyco-biomarkers in serum using newly developed technologies for glycoproteomics. FEBS J. 277, 95–105 (2010).
Qin, X. et al. Comparative analysis for glycopatterns and complex-type N-glycans of glycoprotein in sera from chronic hepatitis B- and C-infected patients. Front. Physiol. 8, 596 (2017).
Yamashita, K. et al. Lectin microarray technology identifies specific lectins related to lymph node metastasis of advanced gastric cancer. Gastric Cancer 19, 531–542 (2016).
Qin, Y. et al. Alteration of liver glycopatterns during cirrhosis and tumor progression induced by HBV. Glycoconj. J. 33, 125–136 (2016).
Takayama, H. et al. Altered glycosylation associated with dedifferentiation of hepatocellular carcinoma: a lectin microarray-based study. BMC Cancer 20, 192 (2020).
Inoue, K. et al. Urinary fetuin-A is a novel marker for diabetic nephropathy in type 2 diabetes identified by lectin microarray. PLoS ONE 8, e77118 (2013).
Zhu, H. et al. Glycopatterns of urinary protein as new potential diagnosis indicators for diabetic nephropathy. J. Diabetes Res. 2017, 5728087 (2017).
Qin, Y. et al. Age- and sex-associated differences in the glycopatterns of human salivary glycoproteins and their roles against influenza A virus. J. Proteome Res. 12, 2742–2754 (2013).
Shu, J. et al. Salivary glycopatterns as potential biomarkers for diagnosis of gastric cancer. Oncotarget 8, 35718–35727 (2017).
Liu, X. et al. Salivary glycopatterns as potential biomarkers for screening of early-stage breast cancer. EBioMedicine 28, 70–79 (2018).
Ebe, Y. et al. Application of lectin microarray to crude samples: differential glycan profiling of lec mutants. J. Biochem. 139, 323–327 (2006).
Hirabayashi, J. Concept, strategy and realization of lectin-based glycan profiling. J. Biochem. 144, 139–147 (2008).
Propheter, D. C., Hsu, K. L. & Mahal, L. K. Recombinant lectin microarrays for glycomic analysis. Methods Mol. Biol. 723, 67–77 (2011).
Ribeiro, J. P. & Mahal, L. K. Dot by dot: analyzing the glycome using lectin microarrays. Curr. Opin. Chem. Biol. 17, 827–831 (2013).
Hu, D., Tateno, H., Kuno, A., Yabe, R. & Hirabayashi, J. Directed evolution of lectins with sugar-binding specificity for 6-sulfo-galactose. J. Biol. Chem. 287, 20313–20320 (2012).
Fahy, E., Sud, M., Cotter, D. & Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 35, W606–W612 (2007).
Rozman, M., Fabris, D., Mrla, T. & Vukelic, Z. Database and data analysis application for structural characterization of gangliosides and sulfated glycosphingolipids by negative ion mass spectrometry. Carbohydr. Res. 400, 1–8 (2014).
Huang, F. T., Han, Y. B., Feng, Y. & Yang, G. Y. A facile method for controlling the reaction equilibrium of sphingolipid ceramide N-deacylase for lyso-glycosphingolipid production. J. Lipid Res. 56, 1836–1842 (2015).
Chen, W. et al. N-glycan profiles in H9N2 avian influenza viruses from chicken eggs and human embryonic lung fibroblast cells. J. Virol. Methods 249, 10–20 (2017).
Liang, Y. et al. Differentially expressed glycosylated patterns of α-1-antitrypsin as serum biomarkers for the diagnosis of lung cancer. Glycobiology 25, 331–340 (2015).
Song, X. et al. Oxidative release of natural glycans for functional glycomics. Nat. Methods 13, 528–534 (2016).
Bisel, B., Pavone, F. S. & Calamai, M. GM1 and GM2 gangliosides: recent developments. Biomol. Concepts 5, 87–93 (2014).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 81372365 and no. 81871955).
Author information
Authors and Affiliations
Contributions
H.D. and H.Y. designed and performed the experiments. H.D., H.Y. and F.Y. designed and performed the data acquisition and analysis. H.D. and Z.L. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Ruijun Tian and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Du, H. et al. Anal. Chem. 91, 10663–10671 (2019): https://doi.org/10.1021/acs.analchem.9b01945
Key data used in this protocol
Du, H. et al. Anal. Chem. 91, 10663–10671 (2019): https://doi.org/10.1021/acs.analchem.9b01945
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Supplementary Tables 1–3.
Source data
Source Data Fig. 2
Statistical source data
Source Data Fig. 3
Statistical source data
Source Data Fig. 4
Statistical source data
Rights and permissions
About this article
Cite this article
Du, H., Yu, H., Yang, F. et al. Comprehensive analysis of glycosphingolipid glycans by lectin microarrays and MALDI-TOF mass spectrometry. Nat Protoc 16, 3470–3491 (2021). https://doi.org/10.1038/s41596-021-00544-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00544-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.