Abstract
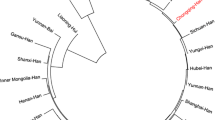
Rwanda is one of the smallest countries of Africa, where forensic genetic studies are rarely being conducted and very few DNA databases have been developed. Short tandem repeats (STRs) polymorphisms were investigated in 505 unrelated Rwandese by using the HUMDNA TYPING (Yanhuang) Kit. The following STRs were targeted: D3S1358, D13S317, D7S820, D16S539, SE33, D10S1248, D5S818, D21S11, TPOX, D1S1656, D6S1043, D19S433, D22S1045, D8S1179, Penta E, D2S441, D12S391, D2S1338, vWA, Penta D, TH01, D18S51, CSF1PO and FGA. The purpose of this study was to elucidate the genetic diversity and explore the potential of applying these 24 STR in 505 Rwandan population in forensics. A total of 360 alleles, with corresponding allele frequencies in the range from 0.001 to 0.442, were found in the Rwandan population. SE33 presented the highest polymorphism (PIC=0.921) among these 24 loci, whereas D13S317 presented the lowest one (PIC=0.671). No deviation from the Hardy-Weinberg equilibrium was observed for any of the 24 loci. The forensic parameters, including the combined power of discrimination (PD and the combined exclusion power, have demonstrated that this panel of 24 STRs is highly informative and useful for forensic applications such as individuals’ identification and paternity tests. Additionally, the genetic distances between Rwanda population and other 24 published populations were calculated based on 8 overlapping loci with the polygenetic tree revealing significant clusters in the populations associated with their geographic locations and their historical relationship.



Similar content being viewed by others
References
Abu-Amero KK, Hellani A, González AM, Larruga JM, Cabrera VM, Underhill PA (2009) Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions. BMC Genet 10(1):59
Agrawal S, Khan F, Talwar S, Nityanand S (2004) Short tandem repeat technology has diverse applications: individual identification, phylogenetic reconstruction and chimerism based post haematopoietic stem cell transplantation graft monitoring. Indian J Med Sci 58(7):297
Alves C, Gusmao L, López-Parra AM, Mesa MS, Amorim A, Arroyo-Pardo E (2005) STR allelic frequencies for an African population sample (Equatorial Guinea) using AmpFlSTR identifiler and Powerplex 16 kits. Forensic Sci Int 148(2–3):239–242
Balamurugan K, Duncan G (2012) Y chromosome STR allelic and haplotype diversity in a Rwanda population from East Central Africa. Leg Med 14(2):105–109
Butler JM (2005) Forensic DNA typing: biology, technology, and genetics of STR markers. Elsevier, Amsterdam
Choi E-J, Park K-W, Lee Y-H, Nam Y-H, Suren G, Ganbold U, Kim J-A, Kim S-Y, Kim H-M, Kim K (2017) Forensic and population genetic analyses of the GlobalFiler STR loci in the Mongolian population. Genes Genom 39(4):423–431
Drobnic K, Pojskic N, Bakal N, Marjanovic D (2005) Allele frequencies for the 15 short tandem repeat loci in Slovenian population. J Forensic Sci 50(6):JFS2005230–JFS2005233
Forward BW, Eastman MW, Nyambo TB, Ballard RE (2008) AMPFlSTR® IdentifilerTM STR allele frequencies in Tanzania Africa. J Forensic Sci 53(1):245–247
Gomes V, Sánchez-Diz P, Alves C, Gomes I, Amorim A, Carracedo Á, Gusmão L (2009) Population data defined by 15 autosomal STR loci in Karamoja population (Uganda) using AmpF/STR Identifiler kit. Forensic Sci Int Genet 3(2):e55–e58
Gymrek M, Willems T, Reich D, Erlich Y (2017) Interpreting short tandem repeat variations in humans using mutational constraint. Nat Genet 49(10):1495
Hammond HA, Jin L, Zhong Y, Caskey CT, Chakraborty R (1994) Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am J Hum Genet 55(1):175
He G, Wang Z, Wang M, Hou Y (2017) Genetic diversity and phylogenetic differentiation of Southwestern Chinese Han: a comprehensive and comparative analysis on 21 non-CODIS STRs. Sci Rep 7(1):13730
Hearne CM, Ghosh S, Todd JA (1992) Microsatellites for linkage analysis of genetic traits. Trends Genet 8(8):288–294
Jeffreys AJ, Tamaki K, MacLeod A, Monckton DG, Neil DL, Armour JA (1994) Complex gene conversion events in germline mutation at human minisatellites. Nat Genet 6(2):136
Louis WR (1963) Ruanda-Urundi. Clarendon Press, Oxford, pp 1884–1919
Mamdani M (2014) When victims become killers: Colonialism, nativism, and the genocide in Rwanda. Princeton University Press, Princeton
Niwenshuti M (2013) Democracy and development in Rwanda?: an assessment of the state of democracy in post-genocide Rwanda and its implications for the Churches' prophetic responsibility. KwaZulu-Natal, Pietermaritzburg
Rakha A, Yu B, Hadi S, Sheng-bin L (2009) Population genetic data on 15 autosomal STRs in a Pakistani population sample. Leg Med 11(6):305–307
Rosenberg LE, Rosenberg DD (2012) Human genes and genomes: science, health, society. Academic Press, Cambridge
Rwanda Population (2021) http://worldpopulationreview.com/countries/rwanda. Accessed 14 May 2021
Semo AC, Carvalho MR, Bogas V, Serra A, Lopes V, Brito P, Sá FB, Porto MJ, Gonçalves IMT, Corte-Real F (2017) Allelic frequencies of 15 autosomal STRs from two main population groups (Makua and Changana) in Mozambique. Forensic Sci Int 6:e286–e288
Slatkin M, Excoffier L (1996) Testing for linkage disequilibrium in genotypic data using the Expectation-Maximization algorithm. Heredity 76(4):377
Sosnov M (2007) The adjudication of genocide: Gacaca and the road to reconciliation in Rwanda. Denv J Int Law Policy 36:125
Teng Y, Zhang F-X, Shen C-M, Wang F, Wang H-D, Yan J-W, Liu J-L (2012) Genetic variation of new 21 autosomal short tandem repeat loci in a Chinese Salar ethnic group. Mol Biol Rep 39(2):1465–1470
Tofanelli S, Boschi I, Bertoneri S, Coia V, Taglioli L, Franceschi MG, Destro-Bisol G, Pascali V, Paoli G (2003) Variation at 16 STR loci in Rwandans (Hutu) and implications on profile frequency estimation in Bantu-speakers. Int J Legal Med 117(2):121–126
Uwineza A, Mutesa L (2015) Medical genetics and genomic medicine in Rwanda. Mol Genet Genom Med 3(6):486–489
Acknowledgements
The authors gratefully thanks to all the Rwanda donors for participating in the present study. The author also would like to thank the Beijing Genomics Institute (BGI-Shenzhen) laboratory for the support during the investigation, also the Rwanda Ministry of Health and of Xi’an Jiaotong University Health Science Center for their cooperation. Ethical clearance was sought from the Ministry of Health-Rwanda. The protocols including sample collections and subsequent analyses were approved by the ethics committee of Xi’an Jiaotong University Health Science Center. All individuals were adequately informed and an appropriate informed consent was obtained before their participation in blood donor by Rwanda Transfusion Centre Laboratory. Before starting our actual sampling, the head of the laboratory approaches participants to inform them about our study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors report no biomedical financial interests or potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Natural Science Foundation of Shaanxi Province (2017JQ8010). Fundamental Research Funds for the Central Universities (xjj2017135). Health Research Fund Project of Shaanxi (2018A010).
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Paul, G., Liu, J., Ma, P. et al. Genetic Polymorphism of 24 Autosomal STR in the Population of Rwanda. Biochem Genet 60, 80–93 (2022). https://doi.org/10.1007/s10528-021-10067-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10067-6