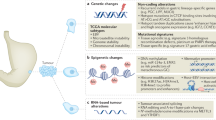
Abstract
There are numerous histological subtypes (histotypes) of gynecological malignancies, with each histotype considered to largely reflect a feature of the “cell of origin,” and to be tightly linked with the clinical behavior and biological phenotype of the tumor. The recent advances in massive parallel sequencing technologies have provided a more complete picture of the range of the genomic alterations that can persist within individual tumors, and have highlighted the types and frequencies of driver-gene mutations and molecular subtypes often associated with these histotypes. Several large-scale genomic cohorts, including the Cancer Genome Atlas (TCGA), have been used to characterize the genomic features of a range of gynecological malignancies, including high-grade serous ovarian carcinoma, uterine corpus endometrial carcinoma, uterine cervical carcinoma, and uterine carcinosarcoma. These datasets have also been pivotal in identifying clinically relevant molecular targets and biomarkers, and in the construction of molecular subtyping schemes. In addition, the recent widespread use of clinical sequencing for the more ubiquitous types of gynecological cancer has manifested in a series of large genomic datasets that have allowed the characterization of the genomes, driver mutations, and histotypes of even rare cancer types, with sufficient statistical power. Here, we review the field of gynecological cancer, and seek to describe the genomic features by histotype. We also will demonstrate how these are linked with clinicopathological attributes and highlight the potential tumorigenic mechanisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15.
Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–94.
Cancer Genome Atlas Research Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73.
Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–5.
Cancer Genome Atlas Research Network, Albert Einstein College of M, Analytical Biological S, Barretos Cancer H, Baylor College of M, Beckman Research Institute of City of H,et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–84.
Scholl S, Popovic M, de la Rochefordiere A, Girard E, Dureau S, Mandic A, et al. Clinical and genetic landscape of treatment naive cervical cancer: alterations in PIK3CA and in epigenetic modulators associated with sub-optimal outcome. EBioMedicine. 2019;43:253–60.
Cherniack AD, Shen H, Walter V, Stewart C, Murray BA, Bowlby R, et al. Integrated molecular characterization of uterine carcinosarcoma. Cancer Cell. 2017;31:411–23.
Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–65 e28.
Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: origin and consequences. Annu Rev Pathol. 2010;5:51–75.
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93.
Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13.
Acharya S, Hensley ML, Montag AC, Fleming GF. Rare uterine cancers. Lancet Oncol. 2005;6:961–71.
Hecht JL, Mutter GL. Molecular and pathologic aspects of endometrial carcinogenesis. J Clin Oncol. 2006;24:4783–91.
Bell DW, Ellenson LH. Molecular genetics of endometrial carcinoma. Annu Rev Pathol. 2019;14:339–67.
Gibson WJ, Hoivik EA, Halle MK, Taylor-Weiner A, Cherniack AD, Berg A, et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet. 2016;48:848–55.
Dou Y, Kawaler EA, Cui Zhou D, Gritsenko MA, Huang C, Blumenberg L, et al. Proteogenomic characterization of endometrial carcinoma. Cell. 2020;180:729–48 e26.
Talhouk A, McConechy MK, Leung S, Li-Chang HH, Kwon JS, Melnyk N, et al. A clinically applicable molecular-based classification for endometrial cancers. Br J Cancer. 2015;113:299–310.
Stelloo E, Nout RA, Osse EM, Jurgenliemk-Schulz IJ, Jobsen JJ, Lutgens LC, et al. Improved risk assessment by integrating molecular and clinicopathological factors in early-stage endometrial cancer-combined analysis of the PORTEC cohorts. Clin Cancer Res. 2016;22:4215–24.
Le Gallo M, Rudd ML, Urick ME, Hansen NF, Zhang S, Program NCS, et al. Somatic mutation profiles of clear cell endometrial tumors revealed by whole exome and targeted gene sequencing. Cancer. 2017;123:3261–8.
DeLair DF, Burke KA, Selenica P, Lim RS, Scott SN, Middha S, et al. The genetic landscape of endometrial clear cell carcinomas. J Pathol. 2017;243:230–41.
Kim SR, Cloutier BT, Leung S, Cochrane D, Britton H, Pina A, et al. Molecular subtypes of clear cell carcinoma of the endometrium: opportunities for prognostic and predictive stratification. Gynecol Oncol. 2020;158:3–11.
Cybulska P, Paula ADC, Tseng J, Leitao MM Jr., Bashashati A, Huntsman DG, et al. Molecular profiling and molecular classification of endometrioid ovarian carcinomas. Gynecol Oncol. 2019;154:516–23.
Kramer P, Talhouk A, Brett MA, Chiu DS, Cairns ES, Scheunhage DA, et al. Endometrial cancer molecular risk stratification is equally prognostic for endometrioid ovarian carcinoma. Clin Cancer Res. 2020;26:5400–10.
Gotoh O, Sugiyama Y, Takazawa Y, Kato K, Tanaka N, Omatsu K, et al. Clinically relevant molecular subtypes and genomic alteration-independent differentiation in gynecologic carcinosarcoma. Nat Commun. 2019;10:4965.
Sugiyama Y, Gotoh O, Fukui N, Tanaka N, Hasumi K, Takazawa Y, et al. Two distinct tumorigenic processes in endometrial endometrioid adenocarcinoma. Am J Pathol. 2020;190:234–51.
Berg A, Hoivik EA, Mjos S, Holst F, Werner HM, Tangen IL, et al. Molecular profiling of endometrial carcinoma precursor, primary and metastatic lesions suggests different targets for treatment in obese compared to non-obese patients. Oncotarget. 2015;6:1327–39.
Russo M, Broach J, Sheldon K, Houser KR, Liu DJ, Kesterson J, et al. Clonal evolution in paired endometrial intraepithelial neoplasia/atypical hyperplasia and endometrioid adenocarcinoma. Hum Pathol. 2017;67:69–77.
Li L, Yue P, Song Q, Yen TT, Asaka S, Wang TL, et al. Genome-wide mutation analysis in precancerous lesions of endometrial carcinoma. J Pathol. 2021;253:119–28.
Zhang L, Kwan SY, Wong KK, Solaman PT, Lu KH, Mok SC. Pathogenesis and clinical management of uterine serous carcinoma. Cancers. 2020;12:686.
Lee EK, Fader AN, Santin AD, Liu JF. Uterine serous carcinoma: molecular features, clinical management, and new and future therapies. Gynecol Oncol. 2021;160:322–32.
Kuhn E, Wu RC, Guan B, Wu G, Zhang J, Wang Y, et al. Identification of molecular pathway aberrations in uterine serous carcinoma by genome-wide analyses. J Natl Cancer Inst. 2012;104:1503–13.
Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–5.
Jones NL, Xiu J, Reddy SK, Burke WM, Tergas AI, Wright JD, et al. Identification of potential therapeutic targets by molecular profiling of 628 cases of uterine serous carcinoma. Gynecol Oncol. 2015;138:620–6.
Wallbillich JJ, Morris RT, Ali-Fehmi R. Comparing mutation frequencies for homologous recombination genes in uterine serous and high-grade serous ovarian carcinomas: a case for homologous recombination deficiency testing in uterine serous carcinoma. Gynecol Oncol. 2020;159:381–6.
Olawaiye AB, Leath CA 3rd. Contemporary management of uterine clear cell carcinoma: a Society of Gynecologic Oncology (SGO) review and recommendation. Gynecol Oncol. 2019;155:365–73.
Baniak N, Fadare O, Kobel M, DeCoteau J, Parkash V, Hecht JL, et al. Targeted molecular and immunohistochemical analyses of endometrial clear cell carcinoma show that POLE mutations and DNA mismatch repair protein deficiencies are uncommon. Am J Surg Pathol. 2019;43:531–7.
Gilks CB, Oliva E, Soslow RA. Poor interobserver reproducibility in the diagnosis of high-grade endometrial carcinoma. Am J Surg Pathol. 2013;37:874–81.
McConechy MK, Ding J, Cheang MC, Wiegand K, Senz J, Tone A, et al. Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol. 2012;228:20–30.
Cuevas D, Valls J, Gatius S, Roman-Canal B, Estaran E, Dorca E, et al. Targeted sequencing with a customized panel to assess histological typing in endometrial carcinoma. Virchows Arch. 2019;474:585–98.
Mhawech-Fauceglia P, Wang D, Kesterson J, Syriac S, Clark K, Frederick PJ, et al. Gene expression profiles in stage I uterine serous carcinoma in comparison to grade 3 and grade 1 stage I endometrioid adenocarcinoma. PLoS ONE. 2011;6:e18066.
Ashley CW, Da Cruz Paula A, Kumar R, Mandelker D, Pei X, Riaz N, et al. Analysis of mutational signatures in primary and metastatic endometrial cancer reveals distinct patterns of DNA repair defects and shifts during tumor progression. Gynecol Oncol. 2019;152:11–9.
Hagemann IS, Deng W, Zaino RJ, Powell MA, Gunderson C, Cosgrove C, et al. The presence of an endometrioid component does not alter the clinicopathologic profile or survival of patients with uterine serous cancer: a gynecologic oncology group (GOG/NRG) study of 934 women. Gynecol Oncol. 2021;160:660–8.
Lawrenson K, Pakzamir E, Liu B, Lee JM, Delgado MK, Duncan K, et al. Molecular analysis of mixed endometrioid and serous adenocarcinoma of the endometrium. PLoS ONE. 2015;10:e0130909.
Espinosa I, D’Angelo E, Palacios J, Prat J. Mixed and ambiguous endometrial carcinomas: a heterogenous group of tumors with different clinicopathologic and molecular genetic features. Am J Surg Pathol. 2016;40:972–81.
Kobel M, Meng B, Hoang LN, Almadani N, Li X, Soslow RA, et al. Molecular analysis of mixed endometrial carcinomas shows clonality in most cases. Am J Surg Pathol. 2016;40:166–80.
Momeni-Boroujeni A, Chiang S. Uterine mesenchymal tumours: recent advances. Histopathology 2020;76:64–75.
McWilliams MM, Chennathukuzhi VM. Recent advances in uterine fibroid etiology. Semin Reprod Med. 2017;35:181–9.
Makinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–5.
Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–10.
Lehtonen R, Kiuru M, Vanharanta S, Sjöberg J, Aaltonen L-M, Aittomäki K, et al. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am J Pathol. 2004;164:17–22.
Quade BJ, Weremowicz S, Neskey DM, Vanni R, Ladd C, Dal Cin P, et al. Fusion transcripts involving HMGA2 are not a common molecular mechanism in uterine leiomyomata with rearrangements in 12q15. Cancer Res. 2003;63:1351–8.
Schoenmakers EF, Bunt J, Hermers L, Schepens M, Merkx G, Janssen B, et al. Identification of CUX1 as the recurrent chromosomal band 7q22 target gene in human uterine leiomyoma. Genes Chromosomes Cancer. 2013;52:11–23.
Mehine M, Kaasinen E, Makinen N, Katainen R, Kampjarvi K, Pitkanen E, et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med. 2013;369:43–53.
Mehine M, Kaasinen E, Heinonen HR, Makinen N, Kampjarvi K, Sarvilinna N, et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc Natl Acad Sci USA. 2016;113:1315–20.
Mehine M, Makinen N, Heinonen HR, Aaltonen LA, Vahteristo P. Genomics of uterine leiomyomas: insights from high-throughput sequencing. Fertil Steril. 2014;102:621–9.
George S, Serrano C, Hensley ML, Ray-Coquard I. Soft tissue and uterine leiomyosarcoma. J Clin Oncol. 2018;36:144–50.
Juhasz-Boss I, Gabriel L, Bohle RM, Horn LC, Solomayer EF, Breitbach GP. Uterine leiomyosarcoma. Oncol Res Treat. 2018;41:680–6.
Mas A, Simon C. Molecular differential diagnosis of uterine leiomyomas and leiomyosarcomas. Biol Reprod. 2019;101:1115–23.
Cuppens T, Moisse M, Depreeuw J, Annibali D, Colas E, Gil-Moreno A, et al. Integrated genome analysis of uterine leiomyosarcoma to identify novel driver genes and targetable pathways. Int J Cancer. 2018;142:1230–43.
Hensley ML, Chavan SS, Solit DB, Murali R, Soslow R, Chiang S, et al. Genomic landscape of uterine sarcomas defined through prospective clinical sequencing. Clin Cancer Res. 2020;26:3881–8.
Choi J, Manzano A, Dong W, Bellone S, Bonazzoli E, Zammataro L, et al. Integrated mutational landscape analysis of uterine leiomyosarcomas. Proc Natl Acad Sci USA. 2021;118:e2025182118.
Kampjarvi K, Makinen N, Kilpivaara O, Arola J, Heinonen HR, Bohm J, et al. Somatic MED12 mutations in uterine leiomyosarcoma and colorectal cancer. Br J Cancer. 2012;107:1761–5.
Williams EA, Sharaf R, Decker B, Werth AJ, Toma H, Montesion M, et al. CDKN2C-Null leiomyosarcoma: a novel, genomically distinct class of TP53/RB1-wild-type tumor with frequent CIC genomic alterations and 1p/19q-codeletion. JCO Precis Oncol. 2020;4:PO.20.00040.
Hoang L, Chiang S, Lee CH. Endometrial stromal sarcomas and related neoplasms: new developments and diagnostic considerations. Pathology. 2018;50:162–77.
Chiang S, Lee CH, Stewart CJR, Oliva E, Hoang LN, Ali RH, et al. BCOR is a robust diagnostic immunohistochemical marker of genetically diverse high-grade endometrial stromal sarcoma, including tumors exhibiting variant morphology. Mod Pathol. 2017;30:1251–61.
Lin DI, Hemmerich A, Edgerly C, Duncan D, Severson EA, Huang RSP, et al. Genomic profiling of BCOR-rearranged uterine sarcomas reveals novel gene fusion partners, frequent CDK4 amplification and CDKN2A loss. Gynecol Oncol. 2020;157:357–66.
Kolin DL, Dong F, Baltay M, Lindeman N, MacConaill L, Nucci MR, et al. SMARCA4-deficient undifferentiated uterine sarcoma (malignant rhabdoid tumor of the uterus): a clinicopathologic entity distinct from undifferentiated carcinoma. Mod Pathol. 2018;31:1442–56.
Lin DI, Allen JM, Hecht JL, Killian JK, Ngo NT, Edgerly C, et al. SMARCA4 inactivation defines a subset of undifferentiated uterine sarcomas with rhabdoid and small cell features and germline mutation association. Mod Pathol. 2019;32:1675–87.
Howitt BE, Sholl LM, Dal Cin P, Jia Y, Yuan L, MacConaill L, et al. Targeted genomic analysis of Mullerian adenosarcoma. J Pathol. 2015;235:37–49.
Lee JC, Lu TP, Changou CA, Liang CW, Huang HN, Lauria A, et al. Genomewide copy number analysis of Mullerian adenosarcoma identified chromosomal instability in the aggressive subgroup. Mod Pathol. 2016;29:1070–82.
Piscuoglio S, Burke KA, Ng CK, Papanastasiou AD, Geyer FC, Macedo GS, et al. Uterine adenosarcomas are mesenchymal neoplasms. J Pathol. 2016;238:381–8.
Hodgson A, Amemiya Y, Seth A, Djordjevic B, Parra-Herran C. High-grade Mullerian adenosarcoma: genomic and clinicopathologic characterization of a distinct neoplasm with prevalent TP53 pathway alterations and aggressive behavior. Am J Surg Pathol. 2017;41:1513–22.
Bean GR, Anderson J, Sangoi AR, Krings G, Garg K. DICER1 mutations are frequent in mullerian adenosarcomas and are independent of rhabdomyosarcomatous differentiation. Mod Pathol. 2019;32:280–9.
Wang Y, Nicholes K, Shih IM. The origin and pathogenesis of endometriosis. Annu Rev Pathol. 2020;15:71–95.
Anglesio MS, Papadopoulos N, Ayhan A, Nazeran TM, Noe M, Horlings HM, et al. Cancer-associated mutations in endometriosis without cancer. N Engl J Med. 2017;376:1835–48.
Suda K, Nakaoka H, Yoshihara K, Ishiguro T, Tamura R, Mori Y, et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep. 2018;24:1777–89.
Lac V, Verhoef L, Aguirre-Hernandez R, Nazeran TM, Tessier-Cloutier B, Praetorius T, et al. Iatrogenic endometriosis harbors somatic cancer-driver mutations. Hum Reprod. 2019;34:69–78.
Lac V, Nazeran TM, Tessier-Cloutier B, Aguirre-Hernandez R, Albert A, Lum A, et al. Oncogenic mutations in histologically normal endometrium: the new normal? J Pathol. 2019;249:173–81.
Moore L, Leongamornlert D, Coorens THH, Sanders MA, Ellis P, Dentro SC, et al. The mutational landscape of normal human endometrial epithelium. Nature. 2020;580:640–6.
Inoue S, Hirota Y, Ueno T, Fukui Y, Yoshida E, Hayashi T, et al. Uterine adenomyosis is an oligoclonal disorder associated with KRAS mutations. Nat Commun. 2019;10:5785.
Kroeger PT Jr., Drapkin R. Pathogenesis and heterogeneity of ovarian cancer. Curr Opin Obstet Gynecol. 2017;29:26–34.
Prat J. New insights into ovarian cancer pathology. Ann Oncol. 2012;23:x111–7.
World Health Organization. Female genital tumours (WHO Classification of Tumours Series, 5th ed., volume 4): International Agency for Research on Cancer; Geneva, Switzerland; 2020.
Fujiwara K, Shintani D, Nishikawa T. Clear-cell carcinoma of the ovary. Ann Oncol. 2016;27:i50–i2.
Chien J, Sicotte H, Fan JB, Humphray S, Cunningham JM, Kalli KR, et al. TP53 mutations, tetraploidy and homologous recombination repair defects in early stage high-grade serous ovarian cancer. Nucleic Acids Res. 2015;43:6945–58.
Hoppe MM, Sundar R, Tan DSP, Jeyasekharan AD. Biomarkers for homologous recombination deficiency in cancer. J Natl Cancer Inst. 2018;110:704–13.
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21.
Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet. 2017;49:1476–86.
Yoshida R, Hagio T, Kaneyasu T, Gotoh O, Osako T, Tanaka N, et al. Pathogenicity assessment of variants for breast cancer susceptibility genes based on BRCAness of tumor sample. Cancer Sci. 2021;112:1310–9.
Etemadmoghadam D, deFazio A, Beroukhim R, Mermel C, George J, Getz G, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15:1417–27.
Wang YK, Bashashati A, Anglesio MS, Cochrane DR, Grewal DS, Ha G, et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat Genet. 2017;49:856–65.
Hu Z, Artibani M, Alsaadi A, Wietek N, Morotti M, Shi T, et al. The repertoire of serous ovarian cancer non-genetic heterogeneity revealed by single-cell sequencing of normal fallopian tube epithelial cells. Cancer Cell. 2020;37:226–42 e7.
Konecny GE, Wang C, Hamidi H, Winterhoff B, Kalli KR, Dering J, et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2014;106:dju249.
Waldron L, Riester M, Birrer M. Molecular subtypes of high-grade serous ovarian cancer: the holy grail? J Natl Cancer Inst. 2014;106:dju297.
Murakami R, Matsumura N, Mandai M, Yoshihara K, Tanabe H, Nakai H, et al. Establishment of a novel histopathological classification of high-grade serous ovarian carcinoma correlated with prognostically distinct gene expression subtypes. Am J Pathol. 2016;186:1103–13.
Tan TZ, Miow QH, Huang RY, Wong MK, Ye J, Lau JA, et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med. 2013;5:1051–66
Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208.
Shih IM, Wang Y, Wang TL. The origin of ovarian cancer species and precancerous landscape. Am J Pathol. 2021;191:26–39.
McDaniel AS, Stall JN, Hovelson DH, Cani AK, Liu CJ, Tomlins SA, et al. Next-generation sequencing of tubal intraepithelial carcinomas. JAMA Oncol. 2015;1:1128–32.
Eckert MA, Pan S, Hernandez KM, Loth RM, Andrade J, Volchenboum SL, et al. Genomics of ovarian cancer progression reveals diverse metastatic trajectories including intraepithelial metastasis to the fallopian tube. Cancer Discov. 2016;6:1342–51.
Labidi-Galy SI, Papp E, Hallberg D, Niknafs N, Adleff V, Noe M, et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat Commun. 2017;8:1093.
Wu RC, Wang P, Lin SF, Zhang M, Song Q, Chu T, et al. Genomic landscape and evolutionary trajectories of ovarian cancer precursor lesions. J Pathol. 2019;248:41–50.
Beirne JP, McArt DG, Roddy A, McDermott C, Ferris J, Buckley NE, et al. Defining the molecular evolution of extrauterine high grade serous carcinoma. Gynecol Oncol. 2019;155:305–17.
Pisanic TR 2nd, Wang Y, Sun H, Considine M, Li L, Wang TH, et al. Methylomic landscapes of ovarian cancer precursor lesions. Clin Cancer Res. 2020;26:6310–20.
Pisanic TR 2nd, Cope LM, Lin SF, Yen TT, Athamanolap P, Asaka R, et al. Methylomic analysis of ovarian cancers identifies tumor-specific alterations readily detectable in early precursor lesions. Clin Cancer Res. 2018;24:6536–47.
Mangili G, Bergamini A, Taccagni G, Gentile C, Panina P, Vigano P, et al. Unraveling the two entities of endometrioid ovarian cancer: a single center clinical experience. Gynecol Oncol. 2012;126:403–7.
Matias-Guiu X, Stewart CJR. Endometriosis-associated ovarian neoplasia. Pathology. 2018;50:190–204.
Pierson WE, Peters PN, Chang MT, Chen LM, Quigley DA, Ashworth A, et al. An integrated molecular profile of endometrioid ovarian cancer. Gynecol Oncol. 2020;157:55–61.
Hollis RL, Thomson JP, Stanley B, Churchman M, Meynert AM, Rye T, et al. Molecular stratification of endometrioid ovarian carcinoma predicts clinical outcome. Nat Commun. 2020;11:4995.
McConechy MK, Ding J, Senz J, Yang W, Melnyk N, Tone AA, et al. Ovarian and endometrial endometrioid carcinomas have distinct CTNNB1 and PTEN mutation profiles. Mod Pathol. 2014;27:128–34.
Anglesio MS, Wang YK, Maassen M, Horlings HM, Bashashati A, Senz J, et al. Synchronous endometrial and ovarian carcinomas: evidence of clonality. J Natl Cancer Inst. 2016;108:djv428.
Schultheis AM, Ng CK, De Filippo MR, Piscuoglio S, Macedo GS, Gatius S, et al. Massively parallel sequencing-based clonality analysis of synchronous endometrioid endometrial and ovarian carcinomas. J Natl Cancer Inst. 2016;108:djv427.
Iida Y, Okamoto A, Hollis RL, Gourley C, Herrington CS. Clear cell carcinoma of the ovary: a clinical and molecular perspective. Int J Gynecol Cancer. 2021;31:605–16.
Arildsen NS, Jonsson JM, Bartuma K, Ebbesson A, Westbom-Fremer S, Masback A, et al. Involvement of chromatin remodeling genes and the Rho GTPases RhoB and CDC42 in ovarian clear cell carcinoma. Front Oncol. 2017;7:109.
Itamochi H, Oishi T, Oumi N, Takeuchi S, Yoshihara K, Mikami M, et al. Whole-genome sequencing revealed novel prognostic biomarkers and promising targets for therapy of ovarian clear cell carcinoma. Br J Cancer. 2017;117:717–24.
Elvin JA, Chura J, Gay LM, Markman M. Comprehensive genomic profiling (CGP) of ovarian clear cell carcinomas (OCCC) identifies clinically relevant genomic alterations (CRGA) and targeted therapy options. Gynecol Oncol Rep. 2017;20:62–6.
Maru Y, Tanaka N, Ohira M, Itami M, Hippo Y, Nagase H. Identification of novel mutations in Japanese ovarian clear cell carcinoma patients using optimized targeted NGS for clinical diagnosis. Gynecol Oncol. 2017;144:377–83.
Murakami R, Matsumura N, Brown JB, Higasa K, Tsutsumi T, Kamada M, et al. Exome sequencing landscape analysis in ovarian clear cell carcinoma shed light on key chromosomal regions and mutation gene networks. Am J Pathol. 2017;187:2246–58.
Shibuya Y, Tokunaga H, Saito S, Shimokawa K, Katsuoka F, Bin L, et al. Identification of somatic genetic alterations in ovarian clear cell carcinoma with next generation sequencing. Genes Chromosomes Cancer. 2018;57:51–60.
Takenaka M, Kobel M, Garsed DW, Fereday S, Pandey A, Etemadmoghadam D, et al. Survival following chemotherapy in ovarian clear cell carcinoma is not associated with pathological misclassification of tumor histotype. Clin Cancer Res. 2019;25:3962–73.
Caumanns JJ, Wisman GBA, Berns K, van der Zee AGJ, de Jong S. ARID1A mutant ovarian clear cell carcinoma: A clear target for synthetic lethal strategies. Biochim Biophys Acta Rev Cancer. 2018;1870:176–84.
Uehara Y, Oda K, Ikeda Y, Koso T, Tsuji S, Yamamoto S, et al. Integrated copy number and expression analysis identifies profiles of whole-arm chromosomal alterations and subgroups with favorable outcome in ovarian clear cell carcinomas. PLoS ONE. 2015;10:e0128066.
Tan TZ, Ye J, Yee CV, Lim D, Ngoi NYL, Tan DSP, et al. Analysis of gene expression signatures identifies prognostic and functionally distinct ovarian clear cell carcinoma subtypes. EBioMedicine. 2019;50:203–10.
Simons M, Simmer F, Bulten J, Ligtenberg MJ, Hollema H, van Vliet S, et al. Two types of primary mucinous ovarian tumors can be distinguished based on their origin. Mod Pathol. 2020;33:722–33.
Perren TJ. Mucinous epithelial ovarian carcinoma. Ann Oncol. 2016;27:i53–i7.
Gorringe KL, Cheasley D, Wakefield MJ, Ryland GL, Allan PE, Alsop K, et al. Therapeutic options for mucinous ovarian carcinoma. Gynecol Oncol. 2020;156:552–60.
Cheasley D, Wakefield MJ, Ryland GL, Allan PE, Alsop K, Amarasinghe KC, et al. The molecular origin and taxonomy of mucinous ovarian carcinoma. Nat Commun. 2019;10:3935.
Mueller JJ, Schlappe BA, Kumar R, Olvera N, Dao F, Abu-Rustum N, et al. Massively parallel sequencing analysis of mucinous ovarian carcinomas: genomic profiling and differential diagnoses. Gynecol Oncol. 2018;150:127–35.
Ryland GL, Hunter SM, Doyle MA, Caramia F, Li J, Rowley SM, et al. Mutational landscape of mucinous ovarian carcinoma and its neoplastic precursors. Genome Med. 2015;7:87.
Mackenzie R, Kommoss S, Winterhoff BJ, Kipp BR, Garcia JJ, Voss J, et al. Targeted deep sequencing of mucinous ovarian tumors reveals multiple overlapping RAS-pathway activating mutations in borderline and cancerous neoplasms. BMC Cancer. 2015;15:415.
Kaldawy A, Segev Y, Lavie O, Auslender R, Sopik V, Narod SA. Low-grade serous ovarian cancer: a review. Gynecol Oncol. 2016;143:433–8.
Jones S, Wang TL, Kurman RJ, Nakayama K, Velculescu VE, Vogelstein B, et al. Low-grade serous carcinomas of the ovary contain very few point mutations. J Pathol. 2012;226:413–20.
Hunter SM, Anglesio MS, Ryland GL, Sharma R, Chiew YE, Rowley SM, et al. Molecular profiling of low grade serous ovarian tumours identifies novel candidate driver genes. Oncotarget. 2015;6:37663–77.
Van Nieuwenhuysen E, Busschaert P, Laenen A, Moerman P, Han SN, Neven P, et al. Loss of 1p36.33 frequent in low-grade serous ovarian cancer. Neoplasia. 2019;21:582–90.
Spreafico A, Oza AM, Clarke BA, Mackay HJ, Shaw P, Butler M, et al. Genotype-matched treatment for patients with advanced type I epithelial ovarian cancer (EOC). Gynecol Oncol. 2017;144:250–5.
Lin DI, Killian JK, Venstrom JM, Ramkissoon SH, Ross JS, Elvin JA. Recurrent urothelial carcinoma-like FGFR3 genomic alterations in malignant Brenner tumors of the ovary. Mod Pathol. 2020;34:983–93.
Tischkowitz M, Huang S, Banerjee S, Hague J, Hendricks WPD, Huntsman DG, et al. Small-cell carcinoma of the ovary, hypercalcemic type-genetics, new treatment targets, and current management guidelines. Clin Cancer Res. 2020;26:3908–17.
Witkowski L, Carrot-Zhang J, Albrecht S, Fahiminiya S, Hamel N, Tomiak E, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46:438–43.
Ramos P, Karnezis AN, Craig DW, Sekulic A, Russell ML, Hendricks WP, et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet. 2014;46:427–9.
Jelinic P, Mueller JJ, Olvera N, Dao F, Scott SN, Shah R, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46:424–6.
Lin DI, Chudnovsky Y, Duggan B, Zajchowski D, Greenbowe J, Ross JS, et al. Comprehensive genomic profiling reveals inactivating SMARCA4 mutations and low tumor mutational burden in small cell carcinoma of the ovary, hypercalcemic-type. Gynecol Oncol. 2017;147:626–33.
Auguste A, Blanc-Durand F, Deloger M, Le Formal A, Bareja R, Wilkes DC, et al. Small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) beyond SMARCA4 mutations: a comprehensive genomic analysis. Cells. 2020;9:1496.
Farkkila A, Haltia UM, Tapper J, McConechy MK, Huntsman DG, Heikinheimo M. Pathogenesis and treatment of adult-type granulosa cell tumor of the ovary. Ann Med. 2017;49:435–47.
Shah SP, Kobel M, Senz J, Morin RD, Clarke BA, Wiegand KC, et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009;360:2719–29.
Roze J, Monroe G, Kutzera J, Groeneweg J, Stelloo E, Paijens S, et al. Whole genome analysis of ovarian granulosa cell tumors reveals tumor heterogeneity and a high-grade TP53-specific subgroup. Cancers. 2020;12:1308.
Alexiadis M, Rowley SM, Chu S, Leung DTH, Stewart CJR, Amarasinghe KC, et al. Mutational landscape of ovarian adult granulosa cell tumors from whole exome and targeted TERT promoter sequencing. Mol Cancer Res. 2019;17:177–85.
Pilsworth JA, Cochrane DR, Neilson SJ, Moussavi BH, Lai D, Munzur AD, et al. Adult-type granulosa cell tumor of the ovary: a FOXL2-centric disease. J Pathol Clin Res. 2021;7:243–52.
Da Cruz Paula A, da Silva EM, Segura SE, Pareja F, Bi R, Selenica P, et al. Genomic profiling of primary and recurrent adult granulosa cell tumors of the ovary. Mod Pathol. 2020;33:1606–17.
Durmus Y, Kilic C, Cakir C, Yuksel D, Boran N, Karalok A, et al. Sertoli-Leydig cell tumor of the ovary: Analysis of a single institution database and review of the literature. J Obstet Gynaecol Res. 2019;45:1311–8.
Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med. 2012;366:234–42.
de Kock L, Terzic T, McCluggage WG, Stewart CJR, Shaw P, Foulkes WD, et al. DICER1 mutations are consistently present in moderately and poorly differentiated sertoli-leydig cell tumors. Am J Surg Pathol. 2017;41:1178–87.
Parikshaa G, Ariba Z, Pranab D, Nalini G, Manish R, Vanita S, et al. Juvenile granulosa cell tumor of the ovary: a comprehensive clinicopathologic analysis of 15 cases. Ann Diagn Pathol. 2021;52:151721.
Kalfa N, Ecochard A, Patte C, Duvillard P, Audran F, Pienkowski C, et al. Activating mutations of the stimulatory g protein in juvenile ovarian granulosa cell tumors: a new prognostic factor? J Clin Endocrinol Metab. 2006;91:1842–7.
Bessiere L, Todeschini AL, Auguste A, Sarnacki S, Flatters D, Legois B, et al. A Hot-spot of in-frame duplications activates the oncoprotein AKT1 in juvenile granulosa cell tumors. EBioMedicine. 2015;2:421–31.
McCluggage WG, Irving JA, Chong AS, Clarke BA, Young RH, Foulkes WD, et al. Ovarian microcystic stromal tumors are characterized by alterations in the beta-catenin-APC pathway and may be an extracolonic manifestation of familial adenomatous polyposis. Am J Surg Pathol. 2018;42:137–9.
Berton-Rigaud D, Devouassoux-Shisheboran M, Ledermann JA, Leitao MM, Powell MA, Poveda A, et al. Gynecologic Cancer InterGroup (GCIG) consensus review for uterine and ovarian carcinosarcoma. Int J Gynecol Cancer. 2014;24:S55–60.
Matsuo K, Takazawa Y, Ross MS, Elishaev E, Podzielinski I, Yunokawa M, et al. Significance of histologic pattern of carcinoma and sarcoma components on survival outcomes of uterine carcinosarcoma. Ann Oncol. 2016;27:1257–66.
Matsuzaki S, Klar M, Matsuzaki S, Roman LD, Sood AK, Matsuo K. Uterine carcinosarcoma: Contemporary clinical summary, molecular updates, and future research opportunity. Gynecol Oncol. 2021;160:586–601.
Jones S, Stransky N, McCord CL, Cerami E, Lagowski J, Kelly D, et al. Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nat Commun. 2014;5:5006.
McConechy MK, Hoang LN, Chui MH, Senz J, Yang W, Rozenberg N, et al. In-depth molecular profiling of the biphasic components of uterine carcinosarcomas. J Pathol Clin Res. 2015;1:173–85.
Zhao S, Bellone S, Lopez S, Thakral D, Schwab C, English DP, et al. Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2016;113:12238–43.
Jones NL, Xiu J, Chatterjee-Paer S, Buckley de Meritens A, Burke WM, Tergas AI, et al. Distinct molecular landscapes between endometrioid and nonendometrioid uterine carcinomas. Int J Cancer. 2017;140:1396–404.
Le Gallo M, Rudd ML, Urick ME, Hansen NF, National Institutes of Health IntramuralSequencing Center Comparative Sequencing P, Merino MJ. et al. The FOXA2 transcription factor is frequently somatically mutated in uterine carcinosarcomas and carcinomas. Cancer. 2018;124:65–73.
Crane E, Naumann W, Tait D, Higgins R, Herzog T, Brown J. Molecular variations in uterine carcinosarcomas identify therapeutic opportunities. Int J Gynecol Cancer. 2020;30:480–4.
Travaglino A, Raffone A, Gencarelli A, Mollo A, Guida M, Insabato L, et al. TCGA classification of endometrial cancer: the place of carcinosarcoma. Pathol Oncol Res. 2020;26:2067–73.
Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8:e191–e203.
Huang J, Qian Z, Gong Y, Wang Y, Guan Y, Han Y, et al. Comprehensive genomic variation profiling of cervical intraepithelial neoplasia and cervical cancer identifies potential targets for cervical cancer early warning. J Med Genet. 2019;56:186–94.
Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47:158–63.
Matsubara A, Sekine S, Ogawa R, Yoshida M, Kasamatsu T, Tsuda H, et al. Lobular endocervical glandular hyperplasia is a neoplastic entity with frequent activating GNAS mutations. Am J Surg Pathol. 2014;38:370–6.
Pietragalla A, Arcieri M, Marchetti C, Scambia G, Fagotti A. Ovarian cancer predisposition beyond BRCA1 and BRCA2 genes. Int J Gynecol Cancer. 2020;30:1803–10.
Garg K, Karnezis AN, Rabban JT. Uncommon hereditary gynaecological tumour syndromes: pathological features in tumours that may predict risk for a germline mutation. Pathology. 2018;50:238–56.
Cerretelli G, Ager A, Arends MJ, Frayling IM. Molecular pathology of Lynch syndrome. J Pathol. 2020;250:518–31.
Buchanan DD, Tan YY, Walsh MD, Clendenning M, Metcalf AM, Ferguson K, et al. Tumor mismatch repair immunohistochemistry and DNA MLH1 methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level germline mismatch repair gene mutation testing. J Clin Oncol. 2014;32:90–100.
Ryan NAJ, Glaire MA, Blake D, Cabrera-Dandy M, Evans DG, Crosbie EJ. The proportion of endometrial cancers associated with Lynch syndrome: a systematic review of the literature and meta-analysis. Genet Med. 2019;21:2167–80.
Kahn RM, Gordhandas S, Maddy BP, Baltich Nelson B, Askin G, Christos PJ, et al. Universal endometrial cancer tumor typing: How much has immunohistochemistry, microsatellite instability, and MLH1 methylation improved the diagnosis of Lynch syndrome across the population? Cancer. 2019;125:3172–83.
Ryan NAJ, McMahon R, Tobi S, Snowsill T, Esquibel S, Wallace AJ, et al. The proportion of endometrial tumours associated with Lynch syndrome (PETALS): a prospective cross-sectional study. PLoS Med. 2020;17:e1003263.
Dominguez-Valentin M, Sampson JR, Seppala TT, Ten Broeke SW, Plazzer JP, Nakken S, et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med. 2020;22:15–25.
Ryan NAJ, Evans DG, Green K, Crosbie EJ. Pathological features and clinical behavior of Lynch syndrome-associated ovarian cancer. Gynecol Oncol. 2017;144:491–5.
Konstantinopoulos PA, Lacchetti C, Annunziata CM. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline summary. JCO Oncol Pract. 2020;16:e835–e8.
Easton DF, Ford D, Bishop DT. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet. 1995;56:265–71.
Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–89.
Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst. 2013;105:812–22.
van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, Hoogerbrugge N, Verhoef S, Vasen HF, et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet. 2005;42:711–9.
Yang X, Leslie G, Doroszuk A, Schneider S, Allen J, Decker B, et al. Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J Clin Oncol. 2020;38:674–85.
Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–82.
Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:18032–7.
Song H, Dicks E, Ramus SJ, Tyrer JP, Intermaggio MP, Hayward J, et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol. 2015;33:2901–7.
Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015;107:djv214.
Norquist BM, Harrell MI, Brady MF, Walsh T, Lee MK, Gulsuner S, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–90.
Pennington KP, Walsh T, Lee M, Pennil C, Novetsky AP, Agnew KJ, et al. BRCA1, TP53, and CHEK2 germline mutations in uterine serous carcinoma. Cancer. 2013;119:332–8.
de Jonge MM, Mooyaart AL, Vreeswijk MP, de Kroon CD, van Wezel T, van Asperen CJ, et al. Linking uterine serous carcinoma to BRCA1/2-associated cancer syndrome: a meta-analysis and case report. Eur J Cancer. 2017;72:215–25.
Long B, Lilyquist J, Weaver A, Hu C, Gnanaolivu R, Lee KY, et al. Cancer susceptibility gene mutations in type I and II endometrial cancer. Gynecol Oncol. 2019;152:20–5.
Sorrell AD, Espenschied CR, Culver JO, Weitzel JN. Tumor protein p53 (TP53) testing and Li-Fraumeni syndrome: current status of clinical applications and future directions. Mol Diagn Ther. 2013;17:31–47.
Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ, et al. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009;27:1250–6.
Mahdi H, Mester JL, Nizialek EA, Ngeow J, Michener C, Eng C. Germline PTEN, SDHB-D, and KLLN alterations in endometrial cancer patients with Cowden and Cowden-like syndromes: an international, multicenter, prospective study. Cancer. 2015;121:688–96.
Tan MH, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res. 2012;18:400–7.
Young RH, Welch WR, Dickersin GR, Scully RE. Ovarian sex cord tumor with annular tubules. Review of 74 cases including 27 with Peutz-Jeghers syndrome and four with adenoma malignum of the cervix. Cancer. 1982;50:1384–402.
Schultz KAP, Williams GM, Kamihara J, Stewart DR, Harris AK, Bauer AJ, et al. DICER1 and associated conditions: identification of at-risk individuals and recommended surveillance strategies. Clin Cancer Res. 2018;24:2251–61.
Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9:eaan2415.
Wang Y, Li L, Douville C, Cohen JD, Yen TT, Kinde I, et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med. 2018;10:eaap8793.
Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci USA. 2012;109:21301–6.
Suva ML, Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell. 2019;75:7–12.
Acknowledgements
The authors thank Rebecca Jackson for editing a draft of this manuscript. This work was supported by JSPS KAKENHI; Grant Numbers JP17K18337, JP15K06861, JP18K07338, JP26462543, and JP17K11308 and by the Vehicle Racing Commemorative Foundation; Grant Numbers 5144, 5274, and 5393.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mori, S., Gotoh, O., Kiyotani, K. et al. Genomic alterations in gynecological malignancies: histotype-associated driver mutations, molecular subtyping schemes, and tumorigenic mechanisms. J Hum Genet 66, 853–868 (2021). https://doi.org/10.1038/s10038-021-00940-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-021-00940-y
This article is cited by
-
Mutational signatures reveal ternary relationships between homologous recombination repair, APOBEC, and mismatch repair in gynecological cancers
Journal of Translational Medicine (2022)