Abstract
A maximum likelihood estimation routine is presented for a generalized structural equation model that permits a combination of response variables from various distributions (e.g., normal, Poisson, binomial, etc.). The likelihood function does not have a closed-form solution and so must be numerically approximated, which can be computationally demanding for models with several latent variables. However, the dimension of numerical integration can be reduced if one or more of the latent variables do not directly affect any nonnormal endogenous variables. The method is demonstrated using an empirical example, and the full estimation details, including first-order derivatives of the likelihood function, are provided.


Similar content being viewed by others
Notes
There are two minor differences between the model implemented here and the model used by Collins et al. (2011). First, Collins et al. (2011) modeled subjective drinking norms using a hierarchical factor model with two sub-factors, corresponding to descriptive and injunctive norms, each measured by two items. Second, Collins et al. (2011) used a quadratic growth model for the number of drinking episodes, whereas a linear growth model is used here.
References
Asparouhov, T., Masyn, K., & Muthen, B. (2006). Continuous time survival in latent variable models. In Proceedings of the joint statistical meeting in Seattle (pp. 180-187).
Bartholomew, D. J., & Knott, M. (1999). Kendall’s library of statistics 7: Latent variable models and factor analysis. London: Edward Arnold.
Bauer, D. J. (2003). Estimating multilevel linear models as structural equation models. Journal of Educational and Behavioral Statistics, 28(2), 135–167.
Bauer, D. J. (2005). A semiparametric approach to modeling nonlinear relations among latent variables. Structural Equation Modeling, 12(4), 513–535.
Bentler, P. M. (2004). Eqs 6: Structural equations program manual. Encino: Multivariate Software.
Bianconcini, S., et al. (2014). Asymptotic properties of adaptive maximum likelihood estimators in latent variable models. Bernoulli, 20(3), 1507–1531.
Cai, L. (2010). High-dimensional exploratory item factor analysis by a Metropolis–Hastings Robbins–Monro algorithm. Psychometrika, 75(1), 33–57.
Collins, S. E., Witkiewitz, K., & Larimer, M. E. (2011). The theory of planned behavior as a predictor of growth in risky college drinking. Journal of Studies on Alcohol and Drugs, 72(2), 322–332.
Curran, P. J. (2003). Have multilevel models been structural equation models all along? Multivariate Behavioral Research, 38(4), 529–569.
du Toit, S. H., & Cudeck, R. (2009). Estimation of the nonlinear random coefficient model when some random effects are separable. Psychometrika, 74(1), 65–82.
Eddelbuettel, D. (2013). Seamless R and C++ integration with Rcpp. New York: Springer. https://doi.org/10.1007/978-1-4614-6868-4.
Eddelbuettel, D., & Sanderson, C. (2014). RcppArmadillo: Accelerating R with high-performance C++ linear algebra. Computational Statistics and Data Analysis, 71, 1054–1063. https://doi.org/10.1016/j.csda.2013.02.005.
Fisher, R. A. (1925). Theory of statistical estimation. In Mathematical proceedings of the Cambridge Philosophical Society (Vol. 22, pp. 700–725).
Fox, J., Nie, Z., & Byrnes, J. (2017). sem: Structural equation models [Computer software manual]. Retrieved from https://CRAN.R-project.org/package=sem (R package version 3.1-9)
Jöreskog, K. G., & Sörbom, D. (1996). Lisrel 8: User’s reference guide. Skokie: Scientific Software International.
Lambert, D. (1992). Zero-in ated Poisson regression, with an application to defects in manufacturing. Technometrics, 34(1), 1–14.
Liu, H. (2007). Growth curve models for zero-in ated count data: An application to smoking behavior. Structural Equation Modeling: A Multidisciplinary Journal, 14(2), 247–279.
Liu, Q., & Pierce, D. A. (1994). A note on Gauss–Hermite quadrature. Biometrika, 81(3), 624–629.
Moustaki, I., & Knott, M. (2000). Generalized latent trait models. Psychometrika, 65(3), 391–411.
Muthén, B. (1984). A general structural equation model with dichotomous, ordered categorical, and continuous latent variable indicators. Psychometrika, 49(1), 115–132.
Muthén, B. O., & Asparouhov, T. (2008). Growth mixture modeling: Analysis with non-Gaussian random effects. In G. Fitzmaurice, M. Davidian, G. Verbeke, & G. Molenberghs (Eds.), Longitudinal data analysis (pp. 143–165). Boca Raton, FL: Chapman & Hall/CRC.
Muthén, B., & Asarouhov, T. (2011). Beyond multilevel regression modeling: Multilevel analysis in a general latent variable framework. In J. Hox & J.K. Roberts (Eds.), Handbook of advanced multilevel analysis (pp. 15-40). New York, NY: Taylor and Francis.
Muthén, B. O. (2002). Beyond SEM: General latent variable modeling. Behaviormetrika, 29(1), 81–117.
Muthén, B. O., Muthén, L. K., & Asparouhov, T. (2017). Regression and mediation analysis using mplus. Los Angeles, CA: Muthén & Muthén.
Muthén, L. K., & Muthén, B. (2017). Mplus version 8 [Computer software manual]. Los Angeles, CA: Muthén & Muthén.
Nash, J. C. (2014). On best practice optimization methods in R. Journal of Statistical Software, 60(2), 1–14.
Neale, M. C., Hunter, M. D., Pritikin, J. N., Zahery, M., Brick, T. R., Kirkpatrick, R. M., et al. (2016). OpenMx 2.0: Extended structural equation and statistical modeling. Psychometrika, 81(2), 535–549.
Pek, J., Sterba, S. K., Kok, B. E., & Bauer, D. J. (2009). Estimating and visualizing nonlinear relations among latent variables: A semiparametric approach. Multivariate Behavioral Research, 44(4), 4070–436.
Pinheiro, J. C., & Bates, D. M. (1995). Approximations to the log-likelihood function in the nonlinear mixed-effects model. Journal of Computational and Graphical Statistics, 4(1), 12–35.
R Core Team. (2019). R: A language and environment for statistical computing [Computer software manual]. Vienna, Austria. Retrieved from https://www.R-project.org/
Rabe-Hesketh, S., & Skrondal, A. (2007). Multilevel and latent variable modeling with composite links and exploded likelihoods. Psychometrika, 72(2), 123.
Rabe-Hesketh, S., Skrondal, A., & Pickles, A. (2002). Reliable estimation of generalized linear mixed models using adaptive quadrature. The Stata Journal, 2(1), 1–21.
Rabe-Hesketh, S., Skrondal, A., & Pickles, A. (2004). Generalized multilevel structural equation modeling. Psychometrika, 69(2), 167–190. https://doi.org/10.1007/bf02295939.
Rabe-Hesketh, S., Skrondal, A., & Pickles, A. (2005). Maximum likelihood estimation of limited and discrete dependent variable models with nested random effects. Journal of Econometrics, 128(2), 301–323.
Rockwood, N. J. (2020). Maximum likelihood estimation of multilevel structural equation models with random slopes for latent covariates. Psychometrika, 85(2), 275–300.
Rosseel, Y. (2012). Lavaan: An R package for structural equation modeling and more. version 0.5–12 (BETA). Journal of Statistical Software, 48(2), 1–36.
Skrondal, A., & Rabe-Hesketh, S. (2003). Multilevel logistic regression for polytomous data and rankings. Psychometrika, 68(2), 267–287.
Skrondal, A., & Rabe-Hesketh, S. (2004). Generalized latent variable modeling: Multilevel, longitudinal, and structural equation models. Boca Raton: CRC Press.
Skrondal, A., & Rabe-Hesketh, S. (2009). Prediction in multilevel generalized linear models. Journal of the Royal Statistical Society: Series A (Statistics in Society), 172(3), 659–687.
StataCorp. (2019a). Stata statistical software: Release 16. StataCorp.
StataCorp. (2019b). Stata structural equation modeling reference manual release 16. StataCorp LLC.
Wang, L. (2010). IRT-ZIP modeling for multivariate zero-in ated count data. Journal of Educational and Behavioral Statistics, 35(6), 671–692.
Zheng, X., & Rabe-Hesketh, S. (2007). Estimating parameters of dichotomous and ordinal item response models with gllamm. The Stata Journal, 7(3), 313–333.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A
In this appendix, the first- and second-order derivatives of the log of the unnormalized conditional posterior distribution of \({\varvec{\eta }}_{1i}\),
with respect to \({\varvec{\eta }}_{1i}\) are provided. These derivatives, which are simply the sum of the derivatives of each of the individual components above, can be used in conjunction with the Newton–Raphson algorithm to obtain the posterior mode and curvature of \(\text {log} \{h^*({\varvec{\eta }}_{1i} | {\mathbf {u}}_i, {\mathbf {y}}_i)\}\) at the mode, which can then be used to center and scale the adaptive quadrature nodes.
First, define the intermediary quantities
which do not depend on \({\varvec{\eta }}_{1i}\) or the data and so only need to be computed once for all individuals (i.e., these quantities remain constant across all i). Using these quantities, the first- and second-order derivatives of the components of \(\text {log}\{ h^*({\varvec{\eta }}_{1i} | {\mathbf {u}}_i, {\mathbf {y}}_i)\}\) with respect to \({\varvec{\eta }}_{1i}\) are
The final two quantities, corresponding to the first- and second-order derivatives of \(\text {log}\{f({\mathbf {u}}_i | {\varvec{\eta }}_{1i})\}\), are the only ones that depend on the distributions of \({\mathbf {u}}_i\). When \(u_{ij}\) is conditionally distributed as a Poisson random variable with mean \(\text {exp}(\xi _{ij})\), then
and
where \({\varvec{\Lambda }}_{u1[j]}\) is the jth row of \({\varvec{\Lambda }}_{u1}\).
Appendix B
Most optimization algorithms use the first-order partial derivatives. As discusses in Section 2.5, the derivatives of the (observed data) log-likelihood can be computed using Equation 35, which is a function of the complete data score function with \({\varvec{\eta }}_{1i}\) set to the quadrature nodes \(\tilde{{\mathbf {t}}}_q\). In this appendix, the derivatives of the complete data log-likelihood with respect to each of the parameter vectors/matrices are provided.
As with the derivatives provided in Appendix A, it helps to first define intermediary quantities that remain constant across all individuals and nodes. Specifically, define:
Next, define the following quantities that do depend on the data and/or quadrature nodes (i.e., \({\varvec{\eta }}_{1i} = \tilde{{\mathbf {t}}}_q\)):
Using these expressions, the first-order derivatives of the complete data log-likelihood with respect to the various parameters that do not depend on the conditional distribution of \({\mathbf {u}}_i\) follow as:
where
The derivatives for \({\varvec{\nu }}_{u}\), \({\varvec{\Lambda }}_{u1}\), and \(\mathbf{K}_u\) depend on \(f({\mathbf {u}}_i | {\varvec{\eta }}_{1i})\) and so will change with different model specifications. In general,
When \(u_{ij}\) is conditionally distributed as a Poisson random variable with mean \(\text {exp}(\xi _{ij})\), then
where \( {\varvec{\nu }}_{u[j]}\), \({\varvec{\Lambda }}_{u1[j]}\), and \(\mathbf{K}_{u[j]}\) are the jth rows of \({\varvec{\nu }}_u\), \({\varvec{\Lambda }}_{u1}\), and \(\mathbf{K}_u\), respectively.
Appendix C
A general method for reparameterizing any specified model to minimize the dimension of numerical integration required is discussed in this section. Recall that, without the dimension reduction technique described in this paper, the dimension of numerical integration required for a generalized SEM is equal to the number of latent factors, m. In this paper, it was demonstrated that the dimension can be reduced from m to \(m_1\), where \(m_1\) is the number of latent factors that directly affect the nonnormal response variables \({\mathbf {u}}_i\). These latent factors can be determined via the columns corresponding to \({\varvec{\Lambda }}_{u1}\) in Equation 10. Specifically, all columns within \({\varvec{\Lambda }}\) for which there is at least one nonzero entry in a row corresponding to \({\varvec{\xi }}_i\) would indicate the latent factors that must be numerically integrated (i.e., \({\varvec{\eta }}_{1i}\)).
In Section 3.2.1, it was demonstrated that the number of factors with nonzero \({\varvec{\Lambda }}\) entries in the rows corresponding to \({\varvec{\xi }}_i\) could be reduced by using a phantom variable for each nonnormal response (or each component of a nonnormal response, such as the zero and count components of a ZIP model). Using a phantom variable for each component of a nonnormal response (i.e., each element of \({\varvec{\xi }}_i\)) would result in a total dimension of integration equal to r. Thus, the maximum number of latent factors that must be numerically integrated is \(\text {min}(m_1, r)\), that is, the minimum of either the original number of latent factors with direct effects on nonnormal responses or the number of phantom variables needed for each of the (components for the) nonnormal responses. In the latent growth factors as outcomes model (Section 3.1.1), the minimum was \(m_1 = 2\) (since \(r = 4\)), whereas in the Poisson regression with latent predictors and overdispersion model (Section 3.2.1), the minimum was \(r = 1\) (since \(m_1 = 5\)).
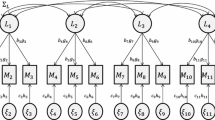
For other models, using a phantom variable for some, but not all, components of nonnormal responses may result in a dimension of integration fewer than both \(m_1\) and r. For example, consider if the indicators for the \(\text {Norm}_i\) factor in the latent regression model with overdispersion displayed in Figure 2 were nonnormal. The original parameterization (top of Figure 2) would require five dimensions of numerical integration, corresponding to each of the latent factors (\(m_1 = 5\)). If a phantom factor was used for all nonnormal response variables (i.e., the four \(\text {Norm}_i\) indicators and \(\text {HED}_{4i}\)), the model would still require five dimensions (\(r = 5\)). If, on the other hand, a phantom factor was only used for \(\text {HED}_{4i}\) (bottom of Figure 2), then the model would only require two dimensions of numerical integration: One for the \(\text {Norm}_i\) factor, as it has direct effects on its nonnormal indicator variables, and one for the \(\text {Disp}_i\) factor, which acts as a phantom factor for \(\text {HED}_{4i}\). Using software such as Mplus, which uses the dimension reduction technique based on \(m_1\) but does not automatically reparameterize the model using phantom variables, the user specifying the model should determine which parameterization would result in the fewest latent variables directly influencing nonnormal responses, so that the dimension of numerical integration required is minimized.
Although determining the proper reparameterization required to minimize the dimension of numerical integration required may seem complex, it is actually possible to determine the correct reparameterization using a basic set of steps:
-
1.
If the original parameterization of a nonnormal response variable already has a single-indicator latent factor (e.g., a dispersion factor), all direct effects of other latent factors on the nonnormal factor should be reparameterized as direct effects on the single-indicator latent factor, which serves as a phantom variable.
-
2.
After any reparameterizations resulting from (1) above, define \({\varvec{\Lambda }}_{u1}^*\) to be a zero-one matrix where an element is zero if the corresponding element in \({\varvec{\Lambda }}_{u1}\) is also zero and one otherwise (i.e., the element in \({\varvec{\Lambda }}_{u1}\) is either a free parameter or a fixed non-zero parameter).
-
3.
Determine each unique row pattern within \({\varvec{\Lambda }}_{u1}^*\).
-
4.
For each unique row pattern within \({\varvec{\Lambda }}_{u1}^*\):
-
(a)
Compute the number of nonnormal components (i.e., elements of \({\varvec{\xi }}_i\)) that have the given row pattern (and denote as \(r^*\)).
-
(b)
Compute the number of latent factors with non-zero effects within the pattern (and denote as \(m^*\)).
-
(a)
-
5.
The latent factors within all patterns in which the number of latent factors is less than or equal to the number of nonnormal components (i.e., \(m^* \le r^*\)) will be numerically integrated.
-
6.
For the remaining patterns, reduce the number of factors to those not included in the list of factors numerically integrated from (5) above (and denote as \(m^{**}\)).
-
7.
For the remaining patterns from (6), if the number of nonnormal components is less than the (reduced) number of latent factors (i.e., \(r^* < m^{**}\)), these nonnormal components should be parameterized using a phantom variable.
To demonstrate the procedure, consider the latent growth factors as outcomes model (Figure 1). Suppose that \(\text {N}_{1i}\) to \(\text {N}_{4i}\) were actually nonnormal responses and \(\text {N}_4\) cross-loaded onto the \(\text {Norm}_i\), \(\text {Att}_i\) and \(\text {Cont}_i\) latent factors. Since there are no single-indicator factors, Step 1 can be skipped. Moving to Step 2,
Following Step 3, the four unique row patterns within \({\varvec{\Lambda }}_{u1}^*\) are determined and displayed in the first five columns of Table 4. The column labeled \(r^*\) contains the number of elements of \({\varvec{\xi }}_i\) (i.e., the number of rows within \({\varvec{\Lambda }}_{u1}^*\)) that is represented by each pattern, and the column labeled \(m^*\) contains the number of latent factors with nonzero elements within the specific row pattern (i.e., Step 4). The second and third rows correspond to patterns with fewer latent factors than nonnormal components (i.e., \(m^* < r^*\)) and so the latent factors with nonzero effects within these patterns will be numerically integrated (Step 5). These include the \(\text {Intercept}_i\) and \(\text {Slope}_i\) factors for the second pattern, and the \(\text {Norm}_i\) factor for the third pattern. The nonzero elements corresponding to these factors are presented in bold within the table.
Next, following Step 6, the remaining number of factors (i.e., non-bold elements) are calculated for each of the remaining patterns (patterns one and four). Since \(\text {Intercept}_i\) and \(\text {Norm}_i\) will already be numerically integrated based on Step 5, the remaining number of factors for patterns one and four is zero and two, respectively. Finally, following Step 7, it is determined that the single nonnormal component within pattern one should not be represented using a phantom factor (since \(r^* > m^{**}\)), whereas the single nonnormal component within pattern four should be reparameterized with a phantom factor (since \(r^* < m^{**}\)). This nonnormal component corresponds to \(N_{4i}\), which is the indicator variable that loads onto the \(\text {Norm}_i\), \(\text {Att}_i\), and \(\text {Cont}_i\) latent factors. The total dimension of numerical integration required for parameter estimation is equal to four, which is the sum of the bold numbers in the final three columns of Table 4. The factors that must be numerically integrated are \(\text {Intercept}_i\), \(\text {Slope}_i\), \(\text {Norm}_i\), and the phantom factor for \(\text {N}_{4i}\).
A major advantage of this approach is that it can be implemented within the estimation routine, rather than requiring the user to conduct the reparameterization by hand before model specification. That is, the original parameterization could be used as input from the user, and the estimation routine could first algorithmically reparameterize the model following the steps outlined above and then estimate the parameters from the reparameterized model, resulting in the dimension of numerical integration required being minimized.
Rights and permissions
About this article
Cite this article
Rockwood, N.J. Efficient Likelihood Estimation of Generalized Structural Equation Models with a Mix of Normal and Nonnormal Responses. Psychometrika 86, 642–667 (2021). https://doi.org/10.1007/s11336-021-09770-5
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11336-021-09770-5