Abstract
Sickle cell disease (SCD) is caused by a mutation in the β-globin gene HBB1. We used a custom adenine base editor (ABE8e-NRCH)2,3 to convert the SCD allele (HBBS) into Makassar β-globin (HBBG), a non-pathogenic variant4,5. Ex vivo delivery of mRNA encoding the base editor with a targeting guide RNA into haematopoietic stem and progenitor cells (HSPCs) from patients with SCD resulted in 80% conversion of HBBS to HBBG. Sixteen weeks after transplantation of edited human HSPCs into immunodeficient mice, the frequency of HBBG was 68% and hypoxia-induced sickling of bone marrow reticulocytes had decreased fivefold, indicating durable gene editing. To assess the physiological effects of HBBS base editing, we delivered ABE8e-NRCH and guide RNA into HSPCs from a humanized SCD mouse6 and then transplanted these cells into irradiated mice. After sixteen weeks, Makassar β-globin represented 79% of β-globin protein in blood, and hypoxia-induced sickling was reduced threefold. Mice that received base-edited HSPCs showed near-normal haematological parameters and reduced splenic pathology compared to mice that received unedited cells. Secondary transplantation of edited bone marrow confirmed that the gene editing was durable in long-term haematopoietic stem cells and showed that HBBS-to-HBBG editing of 20% or more is sufficient for phenotypic rescue. Base editing of human HSPCs avoided the p53 activation and larger deletions that have been observed following Cas9 nuclease treatment. These findings point towards a one-time autologous treatment for SCD that eliminates pathogenic HBBS, generates benign HBBG, and minimizes the undesired consequences of double-strand DNA breaks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
HTS sequencing files can be accessed using the NCBI Sequence Read Archive (PRJNA725249).
Code availability
The code used to conduct off-target quantification and the statistical analysis is available at https://github.com/tsailabSJ/MKSR_off_targets.
References
Piel, F. B., Steinberg, M. H. & Rees, D. C. Sickle cell disease. N. Engl. J. Med. 377, 305 (2017).
Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020).
Miller, S. M. et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 38, 471–481 (2020).
Sangkitporn, S., Rerkamnuaychoke, B., Sangkitporn, S., Mitrakul, C. & Sutivigit, Y. Hb G Makassar (β6:Glu-Ala) in a Thai family. J. Med. Assoc. Thai. 85, 577–582 (2002).
Blackwell, R. Q., Oemijati, S., Pribadi, W., Weng, M. I. & Liu, C. S. Hemoglobin G Makassar: β6 Glu→Ala. Biochim. Biophys. Acta 214, 396–401 (1970).
Wu, L. C. et al. Correction of sickle cell disease by homologous recombination in embryonic stem cells. Blood 108, 1183–1188 (2006).
Leonard, A., Tisdale, J. & Abraham, A. Curative options for sickle cell disease: haploidentical stem cell transplantation or gene therapy? Br. J. Haematol. 189, 408–423 (2020).
Magrin, E., Miccio, A. & Cavazzana, M. Lentiviral and genome-editing strategies for the treatment of β-hemoglobinopathies. Blood 134, 1203–1213 (2019).
Zeng, J. et al. Therapeutic base editing of human hematopoietic stem cells. Nat. Med. 26, 535–541 (2020).
Ribeil, J. A. et al. Gene therapy in a patient with sickle cell disease. N. Engl. J. Med. 376, 848–855 (2017).
Esrick, E. B. et al. Post-transcriptional genetic silencing of BCL11A to treat sickle cell disease. N. Engl. J. Med. 384, 205–215 (2021).
Frangoul, H. et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 384, 252–260 (2021).
Zuccaro, M. V. et al. Allele-specific chromosome removal after Cas9 cleavage in human embryos. Cell 183, 1650–1664.e15 (2020).
Song, Y. et al. Large-fragment deletions induced by Cas9 cleavage while not in the BEs system. Mol. Ther. Nucleic Acids 21, 523–526 (2020).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. & Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930 (2018).
Ihry, R. J. et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018).
Enache, O. M. et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 52, 662–668 (2020).
Wilkinson, A. C. et al. Cas9-AAV6 gene correction of beta-globin in autologous HSCs improves sickle cell disease erythropoiesis in mice. Nat. Commun. 12, 686 (2021).
Dever, D. P. et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389 (2016).
Viprakasit, V., Wiriyasateinkul, A., Sattayasevana, B., Miles, K. L. & Laosombat, V. Hb G-Makassar [β6(A3)Glu→Ala; codon 6 (GAG→GCG)]: molecular characterization, clinical, and hematological effects. Hemoglobin 26, 245–253 (2002).
Chu, S. H. et al. Rationally designed base editors for precise editing of the sickle cell disease mutation. CRISPR J. 4, 169–177 (2021).
Bae, S., Park, J. & Kim, J. S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Tsai, S. Q. et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017).
McIntosh, B. E. et al. Nonirradiated NOD,B6.SCID Il2rγ−/− KitW41/W41 (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Reports 4, 171–180 (2015).
.Alanis-Lobato, G. et al. Frequent loss-of-heterozygosity in CRISPR-Cas9–edited early human embryos. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.2004832117 (2021).
Demirci, S. et al. βT87Q-globin gene therapy reduces sickle hemoglobin production, allowing for ex vivo anti-sickling activity in human erythroid cells. Mol. Ther. Methods Clin. Dev. 17, 912–921 (2020).
Wu, Y. et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med. 25, 776–783 (2019).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Huang, T. P., Newby, G. A. & Liu, D. R. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat. Protoc. 16, 1089–1128 (2021).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Vaidyanathan, S. et al. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol. Ther. Nucleic Acids 12, 530–542 (2018).
Rees, H. A. et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 8, 15790 (2017).
Doman, J. L., Raguram, A., Newby, G. A. & Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628 (2020).
Zuris, J. A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Vakulskas, C. A. et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 24, 1216–1224 (2018).
Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989 (2015).
Connelly, J. P. & Pruett-Miller, S. M. CRIS.py: a versatile and high-throughput analysis program for CRISPR-based genome editing. Sci. Rep. 9, 4194 (2019).
Hu, J. et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood 121, 3246–3253 (2013).
Traxler, E. A. et al. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med. 22, 987–990 (2016).
Lazzarotto, C. R. et al. Defining CRISPR-Cas9 genome-wide nuclease activities with CIRCLE-seq. Nat. Protoc. 13, 2615–2642 (2018).
Hwang, G. H., Kim, J. S. & Bae, S. Web-based CRISPR toolkits: Cas-OFFinder, Cas-Designer, and Cas-Analyzer. Methods Mol. Biol. 2162, 23–33 (2021).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Kumar, S. & Geiger, H. HSC niche biology and HSC expansion ex vivo. Trends Mol. Med. 23, 799–819 (2017).
Leonard, A. et al. Low-dose busulfan reduces human CD34+ cell doses required for engraftment in c-kit mutant immunodeficient mice. Mol. Ther. Methods Clin. Dev. 15, 430–437 (2019).
Karimian, A., Ahmadi, Y. & Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 42, 63–71 (2016).
Kim, H. S., Jeong, Y. K., Hur, J. K., Kim, J. S. & Bae, S. Adenine base editors catalyze cytosine conversions in human cells. Nat. Biotechnol. 37, 1145–1148 (2019).
Acknowledgements
We thank the SCD patients who contributed samples for this study; D. Gao and other members of our laboratories for discussions; and M. O’Reilly for assistance with Adobe Illustrator. This work was supported by US National Institutes of Health awards U01 AI142756, RM1 HG009490, R01 EB031172, R35 GM118062 (D.R.L.), R01R01HL156647 (M.J.W. and D.R.L), U01AI157189 (S.Q.T.), and P01 HL053749 (M.J.W. and S.Q.T.), the Bill and Melinda Gates Foundation (D.R.L.), the Howard Hughes Medical Institute (D.R.L.), the St. Jude Collaborative Research Consortium (D.R.L, S.M.P.-M., S.Q.T and M.J.W.), the Doris Duke Foundation (for aspects of this study that did not use mice; M.J.W., S.Q.T. and A.S.), the American Lebanese Syrian Associated Charities (A.S. and M.J.W.) and the Assisi Foundation of Memphis (M.J.W.). G.A.N. was supported by a Helen Hay Whitney Postdoctoral Fellowship and the HHMI. A.S. was supported by a Scholar Award from the American Society of Hematology. L.W.K. and K.A.E. acknowledge NSF GRFP fellowships. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
G.A.N., J.S.Y., K.J.W., T.M., C.R.L., Y.L., H.S.-T., S.N.P., Y.Y., K.M., K.A.E., Y.J., C.J.P., E.T., C.L., and A.S. conducted experiments and analysed data. J.M.H., M.F.R., K.T.Z., S.M.M., T.W., L.W.K., and J.F.T. prepared materials and provided conceptual assistance, G.A.N., J.S.Y., K.J.W., M.J.W., and D.R.L. wrote the manuscript with input from all authors. J.S.Y., A.P.M., T.A.K., S.Q.T., S.M.P.-M., M.J.W., and D.R.L. supervised this study.
Corresponding authors
Ethics declarations
Competing interests
G.A.N., K.A.E., M.F.R., K.T.Z., S.M.M., T.W., L.W.K., and D.R.L. have filed patent applications on aspects of base editing through the Broad Institute. D.R.L. is a consultant and equity owner of Beam Therapeutics, Prime Medicine, and Pairwise Plants (companies that use genome editing). M.J.W. is on advisory boards for Cellarity Inc., Novartis, and Forma Therapeutics, and is an equity owner of Beam Therapeutics. A.S. is a consultant for Spotlight Therapeutics and his institution receives clinical trial support for the conduct of sickle cell disease gene editing trials from Vertex Pharmaceuticals, CRISPR Therapeutics, and Novartis. J.S.Y. is an equity owner of Beam Therapeutics. The authors declare no non-financial competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Optimization in HEK293T cells, viability and recovery following human SCD HSPC editing, and allelic editing outcomes.
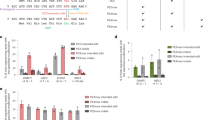
a, Plasmids encoding the HBBS-targeting sgRNA and either ABE7.10-NRCH or ABE8e-NRCH were transfected by lipofection into HEK293T cells. Editing efficiency was measured after 3 days by HTS. Unedited cells were not lipofected. b, Two days after electroporation into human SCD HSPCs of base editor mRNA and sgRNA, or electroporation of ribonucleoprotein (RNP), cell number and viability were measured using a Chemometec Nucleocounter-3000. Acridine orange was used to stain the total cell number and DAPI was used to stain dead, permeabilized cells. The percentage viability was calculated as the DAPI-stained cells divided by the acridine orange cells within each sample. The percentage recovery was normalized to the cell count of the unedited sample. Unedited cells were not electroporated. c, Six days after electroporation of SCD HSPCs, genomic DNA was extracted and the target HBB locus was PCR amplified and sequenced using an Illumina instrument. The sequencing analysis program CRIS.py was used to identify and quantify the resulting alleles. All alleles above a threshold of 0.2% frequency are shown. Below this threshold, variant alleles appeared with greatest frequency in the untreated control sample, suggesting they do not arise from base editor treatment. Nucleotides altered from the endogenous sequence are shown in blue. Rare cytosine base editing was observed at a frequency of less than 1%, as has been previously described as a possible outcome from adenine base editing50. Data shown as mean ± s.d., n = 3.
Extended Data Fig. 2 Erythroid differentiation of edited SCD CD34+ HSPCs.
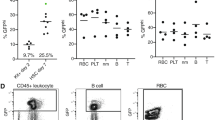
Representative, immuno-flow cytometry for erythroid maturation stage markers42,43 at culture days 7 and 14. Top, gating strategy to identify single cells expressing the erythroid marker hCD235a. Bottom, gating strategy to track the progress of erythroid maturation based on expression of CD49D and BAND3 in hCD235a+ cells. SSC-A, side scatter area; SSC-W, side scatter width; FSC-A, forward scatter area.
Extended Data Fig. 3 Reverse-phase HPLC analysis of erythroid cells derived from in vitro differentiation of edited SCD CD34+ HSPCs.
Reverse-phase HPLC chromatograms of erythroid cell lysates at culture day 18, with β-like globins and their associated fractions marked near the associated peak. Data from the most efficiently edited donor cells are shown. Red arrows indicate the start and end of globin chain peaks.
Extended Data Fig. 4 Off-target base editing associated with ABE8e-NRCH conversion of HBBS to HBBG Makassar in SCD CD34+ HSPCs.
CIRCLE-seq read counts obtained for each verified off-target site and the alignment of each site to the guide sequence are shown. Bar graphs show the percentage of sequencing reads containing A•T-to-G•C mutations within protospacer positions 4–10 at on- and off-target sites in genomic DNA samples from patient CD34+ HSPCs treated with ABE8e-NRCH mRNA, protein, or untreated controls (n = 4). Note that the mutation frequency shown is summed across all reads with one or more A•T-to-G•C mutations in this window. Sequencing errors therefore accumulate in control samples compared to standard sequencing error frequencies for a single nucleotide. Data shown as mean ± s.d.
Extended Data Fig. 5 Off-target indel formation associated with ABE8e-NRCH conversion of HBBS to HBBG Makassar in SCD CD34+ HSPCs.
Bar graph showing the percentage of sequencing reads containing alleles harbouring indels at on- and off-target sites in genomic DNA samples from patient CD34+ HSPCs treated with ABE8e-NRCH mRNA, protein, or untreated controls (n = 4). Data shown as mean ± s.d.
Extended Data Fig. 7 Engraftment of ABE8e-NRCH RNP-treated SCD CD34+ HSPCs after transplantation into immunodeficient mice.
CD34+ HSPCs from three HBBS/S patients with SCD were electroporated with ABE8e-NRCH RNP using an sgRNA targeting the SCD mutant codon, followed by transplantation of 2–5 × 105 treated cells into NBSGW mice via tail-vein injection. Mice were euthanized and analysed 16 weeks after transplantation. a, Experimental workflow. b, Engraftment measured by the percentage of human donor CD45+ cells (hCD45+ cells) in recipient mouse bone marrow. c, Human B cells (hCD19+), myeloid cells (hCD33+), and T cells (hCD3+) in recipient mouse bone marrow, shown as percentages of the total hCD45+ population. d, Human erythroid precursors (hCD235a+) in recipient mouse bone marrow, shown as a percentage of total human and mouse CD45–cells. e, On-target (A7, Fig. 1a) editing efficiencies in human donor CD34+ cell-derived lineages purified from recipient bone marrow by FACS. Erythroid, myeloid, B cell, and HSPC human lineages were collected using antibodies against hCD235a, hCD33, hCD19, and hCD34, respectively. Statistical significance was assessed by one-way ANOVA to compare groups; ns, not significant. f, Percentages of β-like globin proteins determined by reverse-phase HPLC analysis of human donor-derived reticulocytes isolated from recipient mouse bone marrow. g, Representative phase contrast images of human reticulocytes purified from bone marrow and incubated for 8 h with 2% oxygen. Nine images of more than 50 cells per image were collected per sample. Scale bars, 50 μm. h, Quantification of sickled cells calculated by counting images after incubation for 8 h in 2% oxygen as in g. More than 300 randomly selected cells per sample were counted by a blinded observer. n = 14 total mice analysed (b–f); triangle, square, and circle symbols represent samples from three different donors with SCD. Negative control data are shared with Fig. 2. Data shown as mean ± s.d. Statistical significance between treated and untreated samples was assessed using two-tailed Student’s t-test.
Extended Data Fig. 8 Engraftment of transplanted Townes mouse HSPCs, clonality of editing outcomes, and oxygen binding affinity of blood.
a, Donor cell engraftment measured by flow cytometry assessing the percentage of CD45.2+ cells among PBMCs. b, Bone marrow from three mice transplanted with edited Townes mouse HSPCs was plated at low density in methylcellulose. After 12 days of culture, 30 to 35 individual colonies per mouse were picked into cell lysis buffer and the edited locus was amplified by PCR and sequenced by HTS. Colonies were categorized by whether they contained no editing, a monoallelic edit, or a biallelic edit. c, Blood was drawn from mice at week 14 after transplantation. Haemoglobin oxygenation was measured using a Hemox Analyzer (TCS Scientific) across a continuous declining gradient of oxygen pressure to assess whether HBBS-to-HBBG editing led to altered haemoglobin–oxygen binding. Data shown as mean ± s.d.
Extended Data Fig. 9 Adenine base editing of the SCD β-globin allele (HBBS) to the Makassar variant (HBBG) reduces erythrocyte sickling and splenic pathology in mice.
Mice were treated as described in Fig. 3a. Blood and spleen were analysed 16 weeks after transplantation of Lin– mouse HSPCs containing human HBB alleles. a, Representative images of blood smears. One blood smear image was collected per mouse. Scale bars, 25 μm. b, Representative phase contrast images of peripheral blood incubated for 8 h with 2% oxygen. Nine images of more than 50 cells per image were collected per sample. Scale bars, 50 μm. c, Quantification of sickled cells. More than 300 randomly selected cells per condition were counted by a blinded observer. d, Mass of dissected spleens. e, Histological sections of spleens of recipient mice 16 weeks after transplantation. Splenic pathologies in mice that received unedited donor HBBS/S HSCs include excessive extramedullary erythropoiesis and vascular congestion indicated by RBC pooling (bright red colour) resulting in expansion of red pulp (RP), reduction in white pulp (WP), and splenomegaly. Images were taken at 10× magnification and were processed, stained and photographed at the same time under identical conditions. Three images of each spleen were collected from different parts of the organ for each mouse. Scale bars, 100 μm. Unedited HBBS/S, n = 6 mice; edited HBBS/S, n = 6 mice; HBBA/S, n = 2 mice. Data shown as mean ± s.d., with individual values as dots. Statistical significance was assessed using one-way ANOVA with Šidák’s multiple comparisons test of the edited HBBS/S values compared to each other group to calculate P values.
Extended Data Fig. 10 Comparison of DNA damage response and loss of target allele amplification consistent with large deletion or DNA rearrangement in HSPCs following treatment with Cas9 nuclease or with ABE.
HSPCs from a healthy human donor were electroporated in triplicate with Cas9 nuclease RNP targeting the BCL11A erythroid-specific enhancer, ABE8e-NRCH mRNA and an sgRNA targeting the wild-type HBB locus, or no cargo as a control. An additional set of control cells was not electroporated. a, CDKN1 transcription levels, a measure of the p53-mediated DNA damage response49, were quantified by ddPCR after reverse transcription, and were normalized to CDKN1 levels before electroporation (n = 3). b, Editing efficiencies at the targeted genomic loci in HSPCs were measured by HTS 6 days after electroporation. Adenine base editing at the synonymous bystander position 9 of the HBB protospacer is shown for ABE8e-NRCH. c, d, The indicated target sites were amplified and quantified by ddPCR to measure the fraction of missing alleles consistent with larger deletions, translocations, or other chromosomal rearrangements that result in loss of the ability to be amplified by PCR. PCR amplification of a non-targeted ACTB site was used to normalize each sample. Each DNA sample was assessed in triplicate (n = 9). Data shown as mean ± s.d., with individual values in bar graphs shown as dots. Statistical significance between edited and unedited samples was assessed by a two-tailed Student’s t-test; ns, not significant.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Fig. 1, Supplementary Table 1, Supplementary References.
Supplementary Table 2
Off-target sites and primer sequences used for their amplification.
Supplementary Table 3
Complete blood counts from SCD model mice. Complete blood counts were collected from transplanted animals using a FORCYTE veterinary hematology analyzer 16 weeks after the transplantation. Blood counts from untransplanted Townes SCD mice with HBBS/S, HBBA/S, and HBBA/A genotypes were also measured as controls at 4-6 months of age. Data are shown as mean values ±SD.
Rights and permissions
About this article
Cite this article
Newby, G.A., Yen, J.S., Woodard, K.J. et al. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature 595, 295–302 (2021). https://doi.org/10.1038/s41586-021-03609-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03609-w
This article is cited by
-
Deep learning models incorporating endogenous factors beyond DNA sequences improve the prediction accuracy of base editing outcomes
Cell Discovery (2024)
-
An adenine base editor variant expands context compatibility
Nature Biotechnology (2024)
-
Precise genome-editing in human diseases: mechanisms, strategies and applications
Signal Transduction and Targeted Therapy (2024)
-
Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference
Nature Communications (2024)
-
DNA and RNA base editors can correct the majority of pathogenic single nucleotide variants
npj Genomic Medicine (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.