Abstract
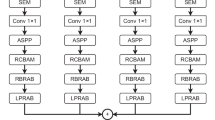
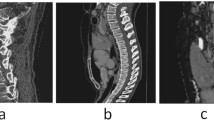
Accurately reading spinal CT images is very important in clinical, but it usually costs some minutes and deeply depends on doctor’s individual experiences. In this paper, we construct a scheme for spinal fracture lesions segmentation based on U-net, by introducing attention module, combining dilated convolution and U-net to get accurate lesions segmentation. First, we present four network schemes to compete in same data set, then get the best one, DU-net(dilated convolution), which replaces original convolution layer with dilated convolution in both contraction path and expansion path of U-net, to increase receptive field for more lesions feature information. Second, we introduce attention module to DU-net for accurate lesions segmentation by focusing on specific regions to improve lesions recognition of training model. Finally, we get prediction results by trained model of lesions segmentation on test data test. The experimental results show that our presented network has a better lesions segmentation performance than U-net, which can save time and reduce patients’ suffering clinically.















Similar content being viewed by others
References
Yu J et al (2019) Hierarchical deep click feature prediction for fine-grained image recognition. IEEE Trans Pattern Anal Mach Intell 99:1–10
Yu J et al (2015) Crossbar-net: a novel convolutional neural network for kidney tumor segmentation in CT images. IEEE Trans Cybern 45(4):767–779
Yu J et al (2020) SPRNet: single-pixel reconstruction for one-stage instance segmentation. IEEE Trans Cybern 99:1–12
Yu Q et al (2019) Crossbar-net: a novel convolutional neural network for kidney tumor segmentation in CT images. IEEE Trans Image Process 99:1–10
Wang Z et al (2017) Hierarchical vertex regression-based segmentation of head and neck CT images for radiotherapy planning. IEEE Trans Image Process 27(2):923–937
Wang S et al (2020) CT male pelvic organ segmentation via hybrid loss network with incomplete annotation. IEEE Trans Medi Imag 99:1
Ronneberger O, Fischer P, Brox T (2015) ’U-Net: Convolutional networks for biomedical image segmentation,’ in Proc. MICCAI, pp:234–241
Mikulka J, et al. (2020) “Pediatric Spine Segmentation and Modeling Using Machine Learning.” 2019 11th International Congress on Ultra Modern Telecommunications and Control Systems and Workshops (ICUMT) IEEE,
Mandal I (2015) Developing new machine learning ensembles for quality spine diagnosis. Knowldege-Based Syst 73:298–310
Tej MS et al (2020) Fostering reproducibility and generalizability in machine learning for clinical prediction modeling in spine surgery. Spine J 45:256
Nam KH, et al. (2019) “Machine Learning Model to Predict Osteoporotic Spine with Hounsfield Units on Lumbar Computed Tomography.” 62
Natalia F, et al. (2019) “Development of Ground Truth Data for Automatic Lumbar Spine MRI Image Segmentation.” 2018 IEEE 20th International Conference on High Performance Computing an Communications; IEEE 16th International Conference on Smart City; IEEE 4th International Conference on Data Science and Systems (HPCC/SmartCity/DSS) IEEE
Khan O et al (2020) Machine learning algorithms for prediction of health-related quality-of-life after surgery for mild degenerative cervical myelopathy. Spine J 56:28
Omar K et al (2020) Prediction of worse functional status after surgery for degenerative cervical myelopathy: a machine learning approach. Neurosurgery 888:584
Lee S et al (2020) The exploration of feature extraction and machine learning for predicting bone density from simple spine X-ray images in a Korean population. Skeletal Radiol 49:613
Shuang Y, et al. (2014) “Feature extraction and classification for ultrasound images of lumbar spine with support vector machine.” IEEE
Wang Z, Zhang Z, Voiculescu I (2020) “RRA-U-Net: a Residual Encoder to Attention Decoder by Residual Connections Framework for Spine Segmentation under Noisy Labels.”
Tang H et al (2020) Automatic lumbar spinal CT image segmentation with a dual densely connected U-Net. IEEE Access 99:1
Li H et al (2021) Automatic lumbar spinal MRI image segmentation with a multi-scale attention network. Neural Comput Appl 10:1–14
Sewon, et al. (2018) “U-net%convolutional neural network%deep learning%fine grain segmentation%intervertebral disc%lumbar spine%magnetic resonance image%segmentation.” Applied sciences (Basel, Switzerland)
Huang J et al (2019) Spine explorer: a deep learning based fully automated program for efficient and reliable quantifications of the vertebrae and discs on sagittal lumbar spine MR images. Spine J 20:4
Korez R et al (2015) A framework for automated spine and vertebrae interpolation-based detection and model-based segmentation. IEEE Trans Med Imag 34(8):1649–1662
Lessmann N, Ginneken BV, Isgum I “Iterative convolutional neural networks for automatic vertebra identification and segmentation in CT images.” SPIE Medical Imaging Conference
Arif Smmra, Knapp K, Slabaugh G (2018) Fully automatic cervical vertebrae segmentation framework for X-ray images. Computer Methods Program Biomed 157:56
Ebrahimi S (2017) “Contribution to automatic adjustments of vertebrae landmarks on x-ray images for 3D reconstruction and quantification of clinical indices.”
Chen Y et al (2019) Vertebrae identification and localization utilizing fully convolutional networks and a hidden Markov model. IEEE Trans Med Imag 99:10
Wang X, Zhai S, Niu Y (2019) Automatic vertebrae localization and identification by combining deep SSAE contextual features and structured regression forest. J Digital Imag 32:336
Li S et al (2020) Multi-task relational learning network for MRI vertebral localization, identification and segmentation. IEEE J Biomed Health Inform 99:1
Shi D, et al. (2018) “Automatic Localization and Segmentation of Vertebral Bodies in 3D CT Volumes with Deep Learning.” the 2nd International Symposium
Lu JT, et al. (2018) “DeepSPINE: Automated Lumbar Vertebral Segmentation, Disc-level Designation, and Spinal Stenosis Grading Using Deep Learning.”
Rak M et al (2019) Combining convolutional neural networks and star convex cuts for fast whole spine vertebra segmentation in MRI. Computer Methods Programs Biomed 177:47–56
Long J, Shelhamer E, Darrell T (2015) “Fully convolutional networks for semantic segmentation,” 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, pp. 3431-3440, https://doi.org/10.1109/CVPR.2015.7298965
Yu F, Koltun V (2016) Multi-Scale Context Aggregation by Dilated Convolutions
Jie H, et al. (2017) “Squeeze-and-Excitation Networks.” IEEE Transactions on Pattern Analysis and Machine Intelligence PP.99
Chuang CH (2019) Efficient triple output network for vertebral segmentation and identification. IEEE Access 7:117978
Acknowledgements
The project in this paper is supported by Biomechanical Modeling of Lumbosacral Spine and Surgical Evaluation System”, Fund Number Nos. 61172147 and 61502365.
Author information
Authors and Affiliations
Contributions
Junsheng Wu contributed to the conception of the study; Gang Sha performed the experiments, contributed significantly to analysis and manuscript preparation and the data analysis and wrote the manuscript; Bin Yu helped to perform the analysis with constructive discussions
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sha, G., Wu, J. & Yu, B. A Robust Segmentation Method Based on Improved U-Net. Neural Process Lett 53, 2947–2965 (2021). https://doi.org/10.1007/s11063-021-10531-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-021-10531-9