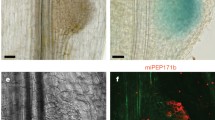
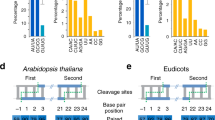
Abstract
It was recently found that the primary transcripts of some microRNA genes (pri-miRNAs) are able to express peptides with 12 to 40 residues in length. These peptides, called miPEPs, participate in the transcriptional regulation of their own pri-miRNAs. In our previous studies, we used bioinformatic approach for comparative analysis of pri-miRNA sequences in plant genomes to identify a new group of miPEPs (miPEP-156a peptides) encoded by pri-miR156a in several dozen species of the Brassicaceae family. Exogenous miPEP-156a peptides could efficiently penetrate into the plant seedlings through the root system and spread systemically to the leaves. The peptides produced moderate morphological effect accelerating primary root growth. In parallel, the miPEP-156a peptides upregulated expression of their own pri-miR156a. Importantly, the observed effects at both morphological and molecular levels correlated with the peptide ability to quickly translocate into the cell nucleus and to bind chromatin. In this work, we established secondary structure of the miPEP-156a and demonstrated its changes induced by formation of the peptide complex with DNA.









Similar content being viewed by others
Notes
* FAM fluorescence detection.
Abbreviations
- CD:
-
circular dichroism
- miRNA:
-
microRNA
- miPEP:
-
peptide encoded by pri-miRNA
- ORF:
-
open reading frame
- pre-miRNA:
-
miRNA precursor
- pri-miRNA:
-
miRNA gene primary transcript
- qPCR:
-
quantitative polymerase chain reaction
- TRAP:
-
translating ribosome affinity immunopurification
References
Wang, J., Mei, J., and Ren, G. (2019) Plant microRNAs: biogenesis, homeostasis, and degradation, Front. Plant Sci., 10, 1-12, https://doi.org/10.3389/fpls.2019.00360.
Bartel, D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function, Cell, 116, 281-297, https://doi.org/10.1016/s0092-8674(04)00045-5.
Rogers, K., and Chen, X. (2013) Biogenesis, turnover, and mode of action of plant microRNAs, Plant Cell, 25, 2383-2399, https://doi.org/10.1105/tpc.113.113159.
Yu, Y., Jia, T., and Chen, X. (2017) The ‘how’ and ‘where’ of plant microRNAs, New Phytol., 216, 1002-1017, https://doi.org/10.1111/nph.14834.
Song, X., Li, Y., Cao, X., and Qi, Y. (2019) MicroRNAs and their regulatory roles in plant–environment interactions, Annu. Rev. Plant Biol., 70, 489-525, https://doi.org/10.1146/annurev-arplant-050718-100334.
Liu, H., Yu, H., Tang, G., and Huang, T. (2018) Small but powerful: function of microRNAs in plant development, Plant Cell Rep., 37, 515-528, https://doi.org/10.1007/s00299-017-2246-5.
Budak, H., and Akpinar, B. A. (2015) Plant miRNAs: biogenesis, organization and origins, Funct. Integr. Genomics, 15, 523-531, https://doi.org/10.1007/s10142-015-0451-2.
Fukudome, A., and Fukuhara, T. (2017) Plant dicer-like proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis, J. Plant Res., 130, 33-44, https://doi.org/10.1007/s10265-016-0877-1.
Zhang, H., Xia, R., Meyers, B. C., and Walbot, V. (2015) Evolution, functions, and mysteries of plant ARGONAUTE proteins, Curr. Opin. Plant Biol., 27, 84-90, https://doi.org/10.1016/j.pbi.2015.06.011.
Tavormina, P., De Coninck, B., Nikonorova, N., De Smet, I., and Cammue, B. P. (2015) The plant peptidome: an expanding repertoire of structural features and biological functions, Plant Cell, 27, 2095-2118, https://doi.org/10.1105/tpc.15.00440.
Zanetti, M. E., Chang, I. F., Gong, F., Galbraith, D. W., and Bailey-Serres, J. (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression, Plant Physiol., 138, 624-635, https://doi.org/10.1104/pp.105.059477.
Juntawong, P., Girke, T., Bazin, J., and Bailey-Serres, J. (2014) Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis, Proc. Natl. Acad. Sci. USA, 111, E203-212, https://doi.org/10.1073/pnas.1317811111.
Zhu, S., Wang, J., He, Y., Meng, N., and Yan, G.-R. (2018) Peptides/proteins encoded by non-coding RNA: a novel resource bank for drug targets and biomarkers, Front. Pharmacol., 9, 1295, https://doi.org/10.3389/fphar.2018.01295.
Hellens, R. P., Brown, C. M., Chisnall, M. A. W., Waterhouse, P. M., and Macknight, R. C. (2016) The emerging world of small ORFs, Trends Plant Sci., 21, 317-328, https://doi.org/10.1016/j.tplants.2015.11.005.
Li, L. J., Leng, R. X., Fan, Y. G., Pan, H. F., and Ye, D. Q. (2017) Translation of noncoding RNAs: focus on lncRNAs, pri-miRNAs, and circRNAs, Exp. Cell Res., 361, 1-8, https://doi.org/10.1016/j.yexcr.2017.10.010.
Chen, Q. J., Deng, B. H., Gao, J., Zhao, Z. Y., Chen, Z. L., et al. (2020) A miRNA-encoded small peptide, vvi-miPEP171d1, regulates adventitious root formation, Plant Physiol., 183, 656-670, https://doi.org/10.1104/pp.20.00197.
Yeasmin, F., Yada, T., and Akimitsu, N. (2018) Micropeptides encoded in transcripts previously identified as long noncoding RNAs: a new chapter in transcriptomics and proteomics, Front. Genet., 9, 144, https://doi.org/10.3389/fgene.2018.00144.
Lauressergues, D., Couzigou, J. M., Clemente, H. S., Martinez, Y., Dunand, C., et al. (2015) Primary transcripts of microRNAs encode regulatory peptides, Nature, 520, 90-93, https://doi.org/10.1038/nature14346.
Couzigou, J. M., André, O., Guillotin, B., Alexandre, M., Combier, J. P. (2016) Use of microRNA-encoded peptide miPEP172c to stimulate nodulation in soybean, New Phytol., 211, 379-381, https://doi.org/10.1111/nph.13991.
Ram, M. K., Mukherjee, K., and Pandey, D. M. (2019) Identification of miRNA, their targets and miPEPs in peanut (Arachishypogaea L.), Comput. Biol. Chem., 83, 107100, https://doi.org/10.1016/j.compbiolchem.2019.107100.
Couzigou, J. M., Lauressergues, D., Bécard, G., and Combier, J. P. (2015) miRNA-encoded peptides (miPEPs): a new tool to analyze the roles of miRNAs in plant biology, RNA Biol., 12, 1178-1180, https://doi.org/10.1080/15476286.2015.1094601.
Morozov, S. Y., Ryazantsev, D. Y., and Erokhina, T. N. (2019) Bioinformatics analysis of the novel conserved micropeptides encoded by the plants of family Brassicaceae, J. Bioinform. Syst. Biol., 2, 066-077, https://doi.org/10.26502/jbsb.5107009.
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis, Plant Physiol., 139, 5-17, https://doi.org/10.1104/pp.105.063743.
Simon, P. (2003) Q-Gene: processing quantitative real-time RT-PCR data, Bioinformatics, 19, 1439-1440, https://doi.org/10.1093/bioinformatics/btg157.
Witus, L. S., and Francis, M. (2010) Site-specific protein bioconjugation via a pyridoxal 5′-phosphate-mediated N-terminal transamination reaction, Curr. Protoc. Chem. Biol., 2, 125-134, https://doi.org/10.1002/9780470559277.ch100018.
Liu, R., and Hu, J. (2013) DNABind: a hybrid algorithm for DNA-binding residue prediction by combining machine learning and template-based approaches, Proteins Struct. Funct. Bioinform., 81, 1888-1899, https://doi.org/10.1002/prot.24330.
Rakitina, D. V., Taliansky, M., Brown, J. W. S., and Kalinina, N. O. (2011) Two RNA-binding sites in plant fibrillarin provide interactions with various RNA substrates, Nucleic Acids Res., 39, 8869-8880, https://doi.org/10.1093/nar/gkr594.
Kim, W., Kim, H. E., Jun, A. R., Jung, M. G., Jin, S., et al. (2016) Structural determinants of miR156a precursor processing in temperature-responsive flowering in Arabidopsis, J. Exp Bot., 67, 4659-4670, https://doi.org/10.1093/jxb/erw248.
Yu, N., Niu, Q. W., Ng, K. H., and Chua, N. H. (2015) The role of miR156/SPLs modules in Arabidopsis lateral root development, Plant J., 83, 673-685, https://doi.org/10.1111/tpj.12919.
Lv, S., Pan, L., and Wang, G. (2016) Commentary: primary transcripts of microRNAs encode regulatory peptides, Front Plant Sci., 22, 1436, https://doi.org/10.3389/fpls.2016.01436.
Shahin-Kaleybar, B., Niazi, A., Afsharifar, A., Nematzadeh, G., Yousefi, R., et al. (2020) Isolation of cysteine-rich peptides from Citrullus colocynthis, Biomolecules, 10, E1326, https://doi.org/10.3390/biom10091326.
Brosnan, C. A., Sarazin, A., Lim, P., Bologna, N. G., Hirsch-Hoffmann, M., and Voinnet, O. (2019) Genome-scale, single-cell-type resolution of microRNA activities within a whole plant organ, EMBO J., 38, e100754, https://doi.org/10.15252/embj.2018100754.
Ormancey, M., Le Ru, A., Duboé, C., Jin, H., Thuleau, P., et al. (2020) Internalization of miPEP165a into Arabidopsis roots depends on both passive diffusion and endocytosis-associated processes, Int. J. Mol. Sci., 21, 2266, https://doi.org/10.3390/ijms21072266.
Sorokin, A. V., Kim, E. R., Ovchinnikov, L. P. (2007) Nucleocytoplasmic transport of proteins, Biochemistry (Moscow), 72, 1439-1457, https://doi.org/10.1134/s0006297907130032.
Merkle, T. (2011) Nucleo-cytoplasmic transport of proteins and RNA in plants, Plant Cell Rep., 30, 153-176, https://doi.org/10.1007/s00299-010-0928-3.
Gizak, A., Maciaszczyk-Dziubinska, E., Jurowicz, M., and Rakus, D. (2009) Muscle FBPase is targeted to nucleus by its 203-KKKGK-207 sequence, Proteins, 77, 262-267, https://doi.org/10.1002/prot.22506.
Lanctot, C., Cheutin, T., Cremer, M., Cavalli, G., and Cremer, T. (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions, Nat. Rev. Genet., 8, 104-115, https://doi.org/10.1038/nrg2041.
Bennetzen, M. V., Larsen, D. H., Bunkenborg, J., Bartek, J., Lukas, J., and Andersen, J. S. (2010) Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response, Mol. Cell. Proteomics, 9, 1314-1323, https://doi.org/10.1074/mcp.M900616-MCP200.
Lai, X., Verhage, L., Hugouvieux, V., and Zubieta, C. (2018) Pioneer factors in animals and plants-colonizing chromatin for gene regulation, Molecules, 23, 1914-1936, https://doi.org/10.3390/molecules23081914.
Lone, I. N., Shukla, M. S., Charles Richard, J. L., Peshev, Z. Y., Dimitrov, S., et al. (2013) Binding of NF-kB to nucleosomes: effect of translational positioning, nucleosome remodeling and linker histone H1, PLoS Genet., 9, e1003830, https://doi.org/10.1371/journal.pgen.1003830.
Horikoshi, N., Kujirai, T., Sato, K., Kimura, H., and Kurumizaka, H. (2019) Structure-based design of an H2A.Z.1 mutant stabilizing a nucleosome in vitro and in vivo, Biochem. Biophys. Res. Commun., 515, 719-724, https://doi.org/10.1016/j.bbrc.2019.06.012.
Van der Lee, R., Buljan, M., Lang, B., Weatheritt, R. J., Daughdrill, G. W., et al. (2014) Classification of intrinsically disordered regions and proteins, Chem. Rev., 114, 6589-6631, https://doi.org/10.1021/cr400525m.
Peng, Z., and Kurgan, L. (2015) High-throughput prediction of RNA, DNA and protein binding regions mediated by intrinsic disorder, Nucleic Acids Res., 43, e121, https://doi.org/10.1093/nar/gkv585.
Wang, C., Uversky, V. N., and Kurgan, L. (2016) Disordered nucleiome: Abundance of intrinsic disorder in the DNA- and RNA-binding proteins in 1121 species from Eukaryota, Bacteria and Archaea, Proteomics, 16, 1486-1498, https://doi.org/10.1002/pmic.201500177.
Sabari, B. R., Dall’Agnese, A., Boija, A., Klein, I. A., Coffey, E. L., et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control, Science, 361, earr3958, https://doi.org/10.1126/science.aar3958.
Quirolo, Z. B., Sequeira, M. A., Martins, J. C., and Dodero, V. I. (2019) Sequence-specific DNA binding by noncovalent peptide – azocyclodextrin dimer complex as a suitable model for conformational fuzziness, Molecules, 24, 2508-2528, https://doi.org/10.3390/molecules24132508.
Miskei, M., Gregus, A., Sharma, R., Duro, N., Zsolyomi, F., and Fuxreiter, M. (2017) Fuzziness enables context dependence protein interactions, FEBS Lett., 591, 2682-2695, https://doi.org/10.1002/1873-3468.12762.
Wright, P. E., and Dyson, H. J. (2009) Linking folding and binding, Curr. Opin. Struct. Biol., 19, 31-38, https://doi.org/10.1016/j.sbi.2008.12.003.
Sharma, A., Badola, P. K., Bhatia, C., Sharma, D., and Trivedi, P. K. (2020) Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis, Nat. Plants, 6, 1262-1274, https://doi.org/10.1038/s41477-020-00769-x.
Acknowledgments
The authors are grateful to A. A. Ignatova (Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry) for help with the CD experiments.
Funding
This work was financially supported by the Russian Foundation for Basic Research (project no. 19-04-00174-a).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflict of interest in financial or any other sphere. The article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Erokhina, T.N., Ryazantsev, D.Y., Samokhvalova, L.V. et al. Activity of Chemically Synthesized Peptide Encoded by the miR156A Precursor and Conserved in the Brassicaceae Family Plants. Biochemistry Moscow 86, 551–562 (2021). https://doi.org/10.1134/S0006297921050047
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297921050047