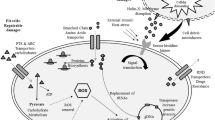
Abstract
Thirteen out of the 15 known Cryobacterium spp. were from extremely cold environments. However, the fundamental question on their cold adaptation strategies to survive in the cold has not been addressed adequately. Hence, this work was conducted to determine the Cryobacterium sp. SO1 cold adaptation strategies. Cryobacterium sp. SO1 that grew optimally at 20 °C was exposed to a sub-optimal temperature of 10 °C. Its mRNA was extracted, sequenced, and analyzed. Strain SO1 global transcriptional profiles revealed a total of 182 differential expressed genes. Four hydrolases, a clp protease, and novel YraN family endonuclease that were related to the programmed cell death pathway were upregulated, indicating that the temperature drop was probably lethal to some cells. Three highly upregulated transcriptional regulators were likely to be the key components to regulate genome-wide expression to adapt to the cold. Meanwhile, the oligo-ribonuclease and a Clp protease gene were upregulated probably to remove accumulated misfolded mRNA and proteins, respectively. The SerB gene was upregulated probably to provide more L-serine residue for the biosynthesis of cold-adapted proteins. Interestingly, most of the stress protein genes in the genome, such as the reactive oxygen species (ROS)-scavenging enzymes were not upregulated. Instead, strain SO1 upregulated the six ribosomal genes which were the target of oxidative nucleobase damage caused by the ROS. This mechanism was probably to ensure that the protein biosynthesis machinery was not affected. Overall, strain SO1 had all the necessary genes and well-coordinated mechanisms to adapt to the sub-optimal growth temperature.



Similar content being viewed by others
References
Allocati N, Masuli M, Ilio CD, De Laurenzi V (2015) Die for the community: an overview of programmed cell death in bacteria. Cell Death Dis 6:e1609
Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53:1331–1341
Amato P, Christner BC (2009) Energy metabolism response to low-temperature and frozen condition in Psychrobacter cryohalolentis. Appl Environ Microbiol 75:711–718
Ander S, Pyl P, Huber W (2014) HTSeq-a python framework to work with high throughput sequencing data. Bioinformatics 31:166–169
Angel R (2012) Total nucleic acid extraction from soil. Protocol Exchange. https://doi.org/10.1038/protex.2012.046
Aranda PS, LaJoie DM, Jorcyk CL (2013) Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis 33:366–369
Auch AF, von Jan M, Klenk H-P, Göker M (2010) Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. SIGS 2:117–134
Bajerski F, Ganzert L, Mangeksdorff K, Lipski A, Wagner D (2011) Cryobacterium arcticum sp. nov., a psychrotolerant bacterium from an Arctic soil. Int J Syst Evol Microbiol 61:1849–1853
Balogh G, Péter M, Glatz A, Gombis I, Török Z, Horváth I, Harwood JL, Vigh L (2013) Key role of lipids in heat stress management. FEBS Lett 587:1970–1980
Bayles KW (2014) Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol 12:63–69
Blagojevic DP, Grubor-Lajsic GN, Spasic MB (2011) Cold defence responses: the role of oxidative stress. Front Biosci 3:416–427
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Bos J, Yakhnina AA, Gitai Z (2012) BapE DNA endonuclease induces an apoptotic-like response to DNA damage in Caulobacter. Proc Natl Sci USA 109:18096–18101
Brinkman AB, Ettema TJG, de Vos WM, van der Oost J (2003) The Lrp family of transcriptional regulators. Mol Microbiol 48:287–294
Chattopadhyay MK (2006) Mechanism of bacterial adaptation to low temperature. J Biosci 31:157–165
Chattopadhyay MK, Jagannadham MV (2001) Maintenance of membrane fluidity in Antarctic bacteria. Polar Biol 24:386–388
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Dastager SG, Lee JC, Ju YJ, Park DJ, Kim CJ (2008) Cryobacterium mesophilum sp. nov., a novel mesophilic bacterium. Int J Syst Evol Microbiol 58:1241–1244
Deng W, Wang H, Xie J (2011) Regulatory and pathogenesis roles of Mycobacterium Lrp/AsnC family transcriptional factors. J Cell Biochem 112:2655–2662
Dieser M, Greenwood A, Foreman CM (2010) Carotenoids pigmentation in Antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct Antarct Alp Res 42:396–405
Duru IC, Ylinen A, Belanov S, Pulido AV, Paulin L, Auvinen P (2021) Transcriptomic time-series analysis of cold- and heat-shock response in psychrotrophic lactic acid bacteria. BMC Genom 22:28
Fogarty CE, Bergmann A (2015) The sound of silence: signaling by apoptotic cells. Curr Top Dev Biol 114:241–265
Fong N, Burgess M, Barrow K, Glenn D (2001) Carotenoids accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl Microbiol Biotechnol 56:750–756
Gao H, Wang Y, Liu X, Yan T, Wu L, Alm E, Arkin A, Thompson DK, Zhou J (2004) Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J Bacteriol 186:7796–7803
Ghosh S, Deutsher MP (1999) Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Sci USA 96:4372–4377
Gong C, Lai Q, Cai H, Jiang Y, Liao H, Liu Y, Xue D (2020) Cryobacterium soli sp. nov., isolated from forest soil. Int J Syst Evol Microbiol 70:675–679
Goordial J, Raymond-Bouchard I, Zolotarov Y, de Bethencourt L, Ronholm J, Shapiro N, Woyke T, Stromvik M, Greer CW, Bakermans C, Whyte L (2016) Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol Ecol 92:fiv154
Goulhen F, Grenier D, Mayrand D (2003) Expression of GroEL and DnaK proteins during the acquisition of a transitory resistance to lethal stresses by Actinobacillus actinomycetemcomitans. Microb Ecol Health Dis 15:120–125
Han L, Lobo S, Reynolds KA (1998) Characterization of β-ketoacyl-acyl carrier protein synthase III from Streptomyces glaucescens and its role in initiation of fatty acid biosynthesis. J Bacteriol 180:4481–4486
Hunt M, De Silva N, Otto TD, Parkhill J, Keana JA, Harris SR (2015) Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294. https://doi.org/10.1186/s13059-015-0849-0
Imlay JA (2015) Diagnosing oxidative stress in bacteria: not as easy as you might think. Curr Opin Microbiol 24:124–131
Inoue K (1976) Quantitative ecology of microorganisms of Syowa station in Antarctica and isolation of pscryophiles. J Gen Appl Microbiol 22:143–150
Jian H, Xiong L, He Y, Xiao X (2015) The regulatory function of LexA is temperature-dependent in the deep-sea bacterium Shewanella peizotolerans WP3. Front Microbiol 6:627
Keto-Timonen R, Hietala N, Palonen E, Hakakorpi A, Lindstrom M, Korkeala H (2016) Cold shock proteins: a minireview with special emphasis Csp-family of enteropathogenic Yersinia. Front Microbiol 7:1151
Kim H, Goo E, Kang Y, Kim J, Hwang I (2012) Regulation of universal stress protein genes by quorum sensing and rpoS in Burkholderia glumae. J Bacteriol 194:982–992
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Koren S, Harhay GP, Smith TPL, Bono JL, Harhay DM, McVey SD, Radune D, Bergman NH, Philippy AM (2013) Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genom Biol 14:R101
Krȕger E, Witt E, Ohlmeier S, Hanschke R, Hecker M (2000) The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J Bacteriol 182:3259–3265
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetic analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kumar S, Suyal DC, Yadav A, Shouche Y, Goel R (2020) Psychrophilic Pseudomonas helmanticensis proteome under simulated cold stress. Cell Stress Chaperones 25:1025–1032
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Horner N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li Y, Kromer B, Schukraft G, Bubenzer O, Huang MR, Wang ZM, Bian LG, Li CS (2014) Growth rate of Usnea aurantiacoatra (Jacq.) Bory on Fildes Peninsula, Antarctica and its climatic background. PLoS ONE 9:e100735
Liu B, Qian SB (2015) Translational reprogramming in stress response. Wiley Interdiscip Rev RNA 5:301–305
Liu Q, Liu H, Wen Y, Zhou Y, Xin Y (2012) Cryobacterium flavum sp. nov., and Cryobacterium luteum sp. nov., isolated from glacier ice. Int J Syst Evol Microbiol 62:1296–1299
Liu Q, Liu H, Zhang J, Zhou Y, Xin Y (2013) Cryobacterium levicorallinum sp. nov., a psychrophilic bacterium isolated from glacier ice. Int J Syst Evol Microbiol 63:2819–2822
Liu Q, Xin YH, Chen XL, Liu HC, Zhou YG, Chen WX (2018) Cryobacterium aureum sp. nov., a psychrophilic bacterium isolated from glacier ice collected from the ice tongue surface. Int J Syst Evol Microbiol 68:1173–1176
Liu Q, Tian JH, Liu HC, Zhou YG, Xin YH (2019a) Cryobacterium melibiosiphilum sp. nov., a psychrophilic bacterium isolated from glacier ice. Int J Syst Evol Microbiol 69:3276–3280
Liu Q, Liu HC, Zhou YG, Xin YH (2019b) Genetic diversity glacier-inhabiting bacteria in China and description of Cryobacterium zongtaii sp. nov. and Arthrobacter glacialis sp. nov. Syst Appl Microbial 42:168–177
Liu Q, Tian JH, Liu HC, Zhou YG, Xin YH (2020) Cryobacterium ruanii sp. nov. and Cryobacterium breve sp. nov., isolated from glaciers. Int J Syst Evol Microbiol 70:1918–1923
Lu J, Holmgren A (2014) The thioredoxin antioxidant system. Free Radic Biol Med 66:75–87
Madan Babu M, Teichmann SA (2003) Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet 19:75–79
Manteca A, Fernandez M, Sanchez J (2006a) Cytological biochemical evidence for an early cell dismantling event in surface cultures of Strepromyces antibioticus. Res Microbiol 157:143–152
Manteca A, Mäder U, Connolly BA, Sanchez J (2006b) A proteomic analysis of Streptomyces coelicolor programmed cell death. Proteomics 6:6008–6022
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:327–331
Metpally RPR, Reddy BVB (2009) Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: insights into the molecular basis of cold adaptation proteins. BMC Genom 10:11
Nagamalleswari E, Rao S, Vasu K, Nagaraja V (2017) Restriction endonuclease triggered bacterial apoptosis as a mechanism for long time survival. Nucleic Acids Res 45:8423–8434
Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M (2020) List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70:5607–5612
Peeters SH, de Jonge MI (2017) For the greater good: Programmed cell death in bacterial communities. Microbiol Res 207:161–169
Peng T, Ma L, Feng X, Tao J, Nan M, Liu Y, Li J, Shen L, Wu X, Yu R, Liu X, Qiu Z, Zeng W (2017) Genomic and transcriptomic analyses reveal adaptation mechanisms of Acidithiobacillus ferrivorans strain Y15 to alpine acid mine drainage. PLoS ONE 12:e0178008
Perkins A, Nelson KJ, Parsonage D, Poole LB, Karplus PA (2015) Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci 40:435–445
Phadtare S (2004) Recent developments in bacterial cold-shock response. Curr Issues Mol Biol 6:125–136
Phadtare S, Inouye M (2008) The cold shock response. EcolSal plus. https://doi.org/10.1128/ecosalplus.5.4.2
Raymond-Bouchard I, Goordial J, Zolotarov Y, Ronholm J, Stromvik M, Bakermans C, Whyte LG (2018) Conserved genomic and amino acid traits of cold adaptation in subzero-growing Arctic permafrost bacteria. FEMS Microbiol Ecol 94:1–14
Reddy GSN, Pradhan S, Manorama R, Shivaji S (2010) Cryobacterium roopkundense sp. nov., a psychrophilic bacterium isolated from glacial soil. Int J Syst Evol Microbiol 60:866–870
Richter M, Roselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Sci USA 106:19126–19131
Romero-Santacreu L, Moreno J, Perez-Ortin J, Alepuz P (2009) Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15:1110–1120
Saunders NF, Thomas T, Curmi PM, Mattick JS, Kuczek E, Slade R, Davis J, Franzmann PD, Boone D, Rusterholtz K, Feldman R, Gates C, Bench S, Sowers K, Kadner K, Aerts A, Dehal P, Detter C, Glavina T, Lucas S, Richardson P, Larimer F, Hauser L, Land M, Cavicchioli R (2003) Mechanisms of thermal adaptation revealed from the genomes of the Antarctic archaea Methanogenium frigidum and Methanococcoides burtonii. Genom Res 13:1580–1588
Savijoki K, Ingmer H, Frees D, Vogensen FK, Palva A, Varmanen P (2003) Heat and DNA damage induction of the LexA-like regulator HdiR from Lactococcus lactis is mediated by RecA and ClpP. Mol Microbiol 50:609–621
Schröder J, Tauch A (2010) Transcriptional regulation of gene expression in Corynebacterium glutamicum: the role of global, master and local regulators in the modular and hierarchical gene regulatory network. FEMS Microbiol Rev 34:685–737
Segal G, Ron EZ (1998) Regulation of heat-shock response in bacteria. Ann NY Acad Sci 851:147–151
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212
Suzuki K-I, Sasaki J, Uramoto M, Nakase T, Komagata K (1997) Cryobacterium psychrophilum gen. nov., sp. nov., nom. rev., comb. nov., an obligated psychophilic actonomycete to accommodate “Curtobacterium psychrophilum” Inoue and Komagata 1976. Int J Syst Evol Microbiol 47:474–478
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG (2002) Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc Natl Acad Sci USA 99:13471–13476
Teoh CP, Wong CMVL, Lee DJH, González MA, Najimudin N, Lee P, Cheah YK (2018) Genome sequences of two cold-adapted Cryobacterium spp. SO1 and SO2 from Fildes Peninsula, Antarctica. Curr Sci 115:1706–1708
Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der Oost J, Rafferty JB (2006) Structural insight into the gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res 34:1439–1449
Travers A (2001) DNA-binding proteins. In: Brenner S, Miller JH (eds) Encyclopedia of genetics. Elsevier Science Inc, Amsterdam, pp 541–544
Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M (1994) Analysis of the induction of general stress protein of Bacillus subtilis. Microbiology 140:741–752
Wang Y, Cao P, Sun P, Zhao J, Sun X, Zhang J, Li C, Xiang W, Wang X (2019) Cryobacterium teoidiphilum sp. nov., isolated from rhizosphere soil of lettuce (var. Ramosa Hort.). Antonie Van Leeuwenhoek 112:1611–1621
Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346
Willi J, Kupfer P, Eveguoz D, Fernandez G, Katz A, Leumann C, Polacek N (2018) Oxidative stress damages rRNA inside the ribosome and differentially affects the catalytic center. Nucleic Acids Res 46:1945–1957
Withers LA, King PJ (1979) Proline: a novel cryoprotectant for the freeze preservation of culture cells of Zea mays L. Plant Physiol 64:675–678
Wolffe AP (1995) The cold-shock response in bacteria. Sci Prog 78:301–310
Wong CMVL, Boo SY, Voo CLY, Zainuddin N, Najimudin N (2019) A comparative transcriptomic analysis provides insights into the cold-adaptation mechanisms of a psychrophilic yeast, Glaciozyma antarctica PI12. Polar Biol 42:541–553
Xue D, Zhang S, Wang C, Gong C (2019) The complete genome sequence and annotation of a psychrophilic Cryobacterium species GCJ02 isolated from cryomorphic soil of a virgin forest. Appl Environ Biotechnol 8:44–48
Yao CL, Somero GN (2012) The impact of acute temperature stress on hemocytes of invasive and native mussels (Mytilus galloprovincialis and Mytilus californianus): DNA damage, membrane integrity, apoptosis and signaling pathways. J Exp Biol 215:4267–4277
Žgur-Bertok D (2013) DNA damage repair and bacterial pathogens. PLoS Pathog 9:e1003711
Zhang DC, Wang HX, Cui HL, Yang Y, Liu HC, Dong XZ, Zhou PJ (2007) Cryobacterium psychrotolerans sp. nov., a novel psychrotolerant bacterium isolate from the China No. 1 glacier. Int J Syst Evol Microbiol 57:866–869
Zhang J, Mao Z, Chong K (2013) A global of uncapped mRNAs under cold stress reveals specific decay patterns and endonucleolytic cleavages in Branchypodium distachyon. Genom Biol 14:R92
Zhu C, Sun B, Liu T, Zheng H, Gu W, Wei H, Sun F, Wang Y, Yang M, Bei W, Peng X, She Q, Xie L, Chen L (2017) Genomic and transcriptomic analyses reveal distinct biological function for cold shock proteins (VpaCspA and VpaCspD) in Vibrio parahaemolyticus CHN25 during low-temperature survival. BMC Genom 18:436
Acknowledgements
The funding support from the Ministry of Science, Technology, and Innovation (MOSTI), Malaysia, under the Antarctica Flagship Programme (Sub-Project 1: FP1213E036) is gratefully acknowledged. We would also like to thank Michelle Wong for reading and amending the manuscript.
Author information
Authors and Affiliations
Contributions
CMVLW and NN conceived the research. TCP designed and conducted experiments. TCP and DJHL performed the data analyses. GMA facilitated the collection of samples for the isolation of bacteria. LPC and CYK provided the guidance to perform some of the experiments. CMVLW and TCP wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Teoh, C.P., Lavin, P., Lee, D.J.H. et al. Genomics and transcriptomics analyses provide insights into the cold adaptation strategies of an Antarctic bacterium, Cryobacterium sp. SO1. Polar Biol 44, 1305–1319 (2021). https://doi.org/10.1007/s00300-021-02883-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-021-02883-8