Abstract
Mitochondrial DNA (mtDNA) variation in common diseases has been underexplored, partly due to a lack of genotype calling and quality-control procedures. Developing an at-scale workflow for mtDNA variant analyses, we show correlations between nuclear and mitochondrial genomic structures within subpopulations of Great Britain and establish a UK Biobank reference atlas of mtDNA–phenotype associations. A total of 260 mtDNA–phenotype associations were new (P < 1 × 10−5), including rs2853822/m.8655 C>T (MT-ATP6) with type 2 diabetes, rs878966690/m.13117 A>G (MT-ND5) with multiple sclerosis, 6 mtDNA associations with adult height, 24 mtDNA associations with 2 liver biomarkers and 16 mtDNA associations with parameters of renal function. Rare-variant gene-based tests implicated complex I genes modulating mean corpuscular volume and mean corpuscular hemoglobin. Seven traits had both rare and common mtDNA associations, where rare variants tended to have larger effects than common variants. Our work illustrates the value of studying mtDNA variants in common complex diseases and lays foundations for future large-scale mtDNA association studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Full summary statistics are provided at https://app.box.com/s/vu9ewgd9sv7ga3ua0lsk0cql444xm3ht/ and Zenodo (https://doi.org/10.5281/zenodo.4609973).
We used data from the following publicly available databases: https://www.mitomap.org/; https://www.ncbi.nlm.nih.gov/clinvar/; https://www.internationalgenome.org/.
Code availability
Code used to process UKBB data and source data used to generate the main figures are available at: https://github.com/clody23/UKBiobank_mtPheWas/.
Source data for Figs. 1–4 are available from: https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/Figure1/, https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/Figure2/ and https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/Figure3_and_4/.
Source data for Extended Data Figs. 1–3 are available from: https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/EDF1/, https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/EDF2/, and https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/EDF3/.
References
Saraste, M. Oxidative phosphorylation at the fin de siècle. Science 283, 1488–1493 (1999).
Giles, R. E., Blanc, H., Cann, H. M. & Wallace, D. C. Maternal inheritance of human mitochondrial DNA. Proc. Natl Acad. Sci. USA 77, 6715–6719 (1980).
Elson, J. L. et al. Analysis of European mtDNAs for recombination. Am. J. Hum. Genet. 68, 145–153 (2001).
Wallace, D. C. Mitochondrial DNA sequence variation in human evolution and disease. Proc. Natl Acad. Sci. USA 91, 8739–8746 (1994).
Wallace, D. C., Brown, M. D. & Lott, M. T. Mitochondrial DNA variation in human evolution and disease. Gene 238, 211–230 (1999).
Elson, J. L., Majamaa, K., Howell, N. & Chinnery, P. F. Associating mitochondrial DNA variation with complex traits. Am. J. Hum. Genet. 80, 378–382 (2007).
Poulton, J. et al. Type 2 diabetes is associated with a common mitochondrial variant: evidence from a population-based case–control study. Hum. Mol. Genet. 11, 1581–1583 (2002).
Wallace, D. C. & Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 5, a021220 (2013).
Keogh, M. J. & Chinnery, P. F. Mitochondrial DNA mutations in neurodegeneration. Biochim. Biophys. Acta 1847, 1401–1411 (2015).
Herrnstadt, C. & Howell, N. An evolutionary perspective on pathogenic mtDNA mutations: haplogroup associations of clinical disorders. Mitochondrion 4, 791–798 (2004).
Samuels, D. C., Carothers, A. D., Horton, R. & Chinnery, P. F. The power to detect disease associations with mitochondrial DNA haplogroups. Am. J. Hum. Genet. 78, 713–720 (2006).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Laurie, C. C. et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 34, 591–602 (2010).
Zhao, S. et al. Strategies for processing and quality control of Illumina genotyping arrays. Brief. Bioinform. 19, 765–775 (2017).
Yamamoto, K. et al. Genetic and phenotypic landscape of the mitochondrial genome in the Japanese population. Commun. Biol. 3, 104 (2020).
Hudson, G., Gomez-Duran, A., Wilson, I. J. & Chinnery, P. F. Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet. 10, e1004369 (2014).
Kozin, M. S. et al. Variants of mitochondrial genome and risk of multiple sclerosis development in russians. Acta Naturae 10, 79–86 (2018).
Tranah, G. J. et al. Mitochondrial DNA sequence variation in multiple sclerosis. Neurology 85, 325–330 (2015).
Preste, R., Vitale, O., Clima, R., Gasparre, G. & Attimonelli, M. HmtVar: a new resource for human mitochondrial variations and pathogenicity data. Nucleic Acids Res. 47, D1202–D1210 (2019).
Mitchell, S. L. et al. Investigating the relationship between mitochondrial genetic variation and cardiovascular-related traits to develop a framework for mitochondrial phenome-wide association studies. BioData Min. 7, 6 (2014).
el-Schahawi, M. et al. Two large Spanish pedigrees with nonsyndromic sensorineural deafness and the mtDNA mutation at nt 1555 in the 12s rRNA gene: evidence of heteroplasmy. Neurology 48, 453–456 (1997).
Casano, R. A. et al. Hearing loss due to the mitochondrial A1555G mutation in Italian families. Am. J. Med. Genet. 79, 388–391 (1998).
Bravo, O., Ballana, E. & Estivill, X. Cochlear alterations in deaf and unaffected subjects carrying the deafness-associated A1555G mutation in the mitochondrial 12S rRNA gene. Biochem. Biophys. Res. Commun. 344, 511–516 (2006).
Yengo, L. et al. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018).
Boal, R. L. et al. Height as a clinical biomarker of disease burden in adult mitochondrial disease. J. Clin. Endocrinol. Metab. 104, 2057–2066 (2019).
Holzer, T. et al. Respiratory chain inactivation links cartilage-mediated growth retardation to mitochondrial diseases. J. Cell Biol. 218, 1853–1870 (2019).
Gómez-Durán, A. et al. Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of Leber’s hereditary optic neuropathy. Biochim. Biophys. Acta 1822, 1216–1222 (2012).
Gómez-Durán, A. et al. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum. Mol. Genet. 19, 3343–3353 (2010).
Chen, A., Raule, N., Chomyn, A. & Attardi, G. Decreased reactive oxygen species production in cells with mitochondrial haplogroups associated with longevity. PLoS ONE 7, e46473 (2012).
Niemi, A.-K. et al. A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur. J. Hum. Genet. 13, 166–170 (2005).
Zhang, J. et al. Strikingly higher frequency in centenarians and twins of mtDNA mutation causing remodeling of replication origin in leukocytes. Proc. Natl Acad. Sci. USA 100, 1116–1121 (2003).
Niemi, A.-K. et al. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum. Genet. 112, 29–33 (2003).
Santoro, A. et al. Mitochondrial DNA involvement in human longevity. Biochim. Biophys. Acta 1757, 1388–1399 (2006).
Dato, S. et al. Association of the mitochondrial DNA haplogroup J with longevity is population specific. Eur. J. Hum. Genet. 12, 1080–1082 (2004).
De Benedictis, G. et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 13, 1532–1536 (1999).
Rose, G. et al. Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians. Eur. J. Hum. Genet. 9, 701–707 (2001).
Pacheu-Grau, D. et al. Mitochondrial antibiograms in personalized medicine. Hum. Mol. Genet. 22, 1132–1139 (2013).
Jurkute, N. & Yu-Wai-Man, P. Leber hereditary optic neuropathy: bridging the translational gap. Curr. Opin. Ophthalmol. 28, 403–409 (2017).
Yu-Wai-Man, P., Turnbull, D. M. & Chinnery, P. F. Leber hereditary optic neuropathy. J. Med. Genet. 39, 162–169 (2002).
Kogelnik, A. M., Lott, M. T., Brown, M. D., Navathe, S. B. & Wallace, D. C. MITOMAP: a human mitochondrial genome database. Nucleic Acids Res. 24, 177–179 (1996).
Chinnery, P. F. & Gomez-Duran, A. Oldies but Goldies mtDNA population variants and neurodegenerative diseases. Front. Neurosci. 12, 682 (2018).
Elliott, H. R., Samuels, D. C., Eden, J. A., Relton, C. L. & Chinnery, P. F. Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet 83, 254–260 (2008).
Achilli, A. et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am. J. Hum. Genet. 75, 910–918 (2004).
Patergnani, S. et al. Mitochondria in multiple sclerosis: molecular mechanisms of pathogenesis. Int. Rev. Cell Mol. Biol. 328, 49–103 (2017).
Achilli, A. et al. Mitochondrial DNA backgrounds might modulate diabetes complications rather than T2DM as a whole. PLoS ONE 6, e21029 (2011).
Navas-Madroñal, M. et al. Enhanced endoplasmic reticulum and mitochondrial stress in abdominal aortic aneurysm. Clin. Sci. 133, 1421–1438 (2019).
Hallac, A., Keshava, H. B., Morris-Stiff, G. & Ibrahim, S. Sigmoid volvulus in a patient with mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS): a rare occurrence. BMJ Case Rep. bcr2015213718 (2016).
Yu-Wai-Man, P. & Newman, N. J. Inherited eye-related disorders due to mitochondrial dysfunction. Hum. Mol. Genet. 26, R12–R20 (2017).
Compston, A. & Coles, A. Multiple sclerosis. Lancet 359, 1221–1231 (2002).
Carstens, P.-O. et al. X-linked myotubular myopathy and recurrent spontaneous pneumothorax: a new phenotype? Neurol. Genet. 5, e327 (2019).
Martín-Hernández, E. et al. Renal pathology in children with mitochondrial diseases. Pediatr. Nephrol. 20, 1299–1305 (2005).
Stewart, J. B. & Chinnery, P. F. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 16, 530–542 (2015).
Degli Esposti, D. et al. Mitochondrial roles and cytoprotection in chronic liver injury. Biochem. Res. Int. 2012, 387626 (2012).
Houten, S. M. & Wanders, R. J. A. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis. 33, 469–477 (2010).
Owen, O. E., Kalhan, S. C. & Hanson, R. W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 277, 30409–30412 (2002).
Pesi, R., Balestri, F. & Ipata, P. L. Metabolic interaction between urea cycle and citric acid cycle shunt: a guided approach. Biochem. Mol. Biol. Educ. 46, 182–185 (2018).
Martínez-Reyes, I. & Chandel, N. S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11, 102 (2020).
Bhargava, P. & Schnellmann, R. G. Mitochondrial energetics in the kidney. Nat. Rev. Nephrol. 13, 629–646 (2017).
Connor, T. M. et al. Mutations in mitochondrial DNA causing tubulointerstitial kidney disease. PLoS Genet. 13, e1006620 (2017).
Galvan, D. L., Green, N. H. & Danesh, F. R. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 92, 1051–1057 (2017).
Hunt, N. J., Kang, S. W. S., Lockwood, G. P., Le Couteur, D. G. & Cogger, V. C. Hallmarks of aging in the liver. Comput. Struct. Biotechnol. J. 17, 1151–1161 (2019).
Mansouri, A., Gattolliat, C.-H. & Asselah, T. Mitochondrial dysfunction and signaling in chronic liver diseases. Gastroenterology 155, 629–647 (2018).
Lee, W. S. & Sokol, R. J. Liver disease in mitochondrial disorders. Semin. Liver Dis. 27, 259–273 (2007).
O’Toole, J. F. Renal manifestations of genetic mitochondrial disease. Int. J. Nephrol. Renovasc. Dis. 7, 57–67 (2014).
Eirin, A., Lerman, A. & Lerman, L. O. The emerging role of mitochondrial targeting in kidney disease. Handb. Exp. Pharmacol. 240, 229–250 (2017).
Moreno-Loshuertos, R. et al. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 38, 1261–1268 (2006).
Correa, C. C., Aw, W. C., Melvin, R. G., Pichaud, N. & Ballard, J. W. O. Mitochondrial DNA variants influence mitochondrial bioenergetics in Drosophila melanogaster. Mitochondrion 12, 459–464 (2012).
Ji, F. et al. Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc. Natl Acad. Sci. USA 109, 7391–7396 (2012).
Bellizzi, D., D’Aquila, P., Giordano, M., Montesanto, A. & Passarino, G. Global DNA methylation levels are modulated by mitochondrial DNA variants. Epigenomics 4, 17–27 (2012).
Fernández-Moreno, M. et al. Mitochondrial DNA haplogroups influence the risk of incident knee osteoarthritis in OAI and CHECK cohorts. A meta-analysis and functional study. Ann. Rheum. Dis. 76, 1114–1122 (2017).
Kazuno, A. et al. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2, e128 (2006).
Suissa, S. et al. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 5, e1000474 (2009).
Salminen, T. S. et al. Mitochondrial genotype modulates mtDNA copy number and organismal phenotype in Drosophila. Mitochondrion 34, 75–83 (2017).
Picard, M. et al. Progressive increase in mtDNA 3243A > G heteroplasmy causes abrupt transcriptional reprogramming. Proc. Natl Acad. Sci. USA 111, E4033–E4042 (2014).
Mottis, A., Herzig, S. & Auwerx, J. Mitocellular communication: shaping health and disease. Science 366, 827–832 (2019).
Fang, H. et al. mtDNA haplogroup N9a increases the risk of type 2 diabetes by altering mitochondrial function and intracellular mitochondrial signals. Diabetes 67, 1441–1453 (2018).
D’Aquila, P., Rose, G., Panno, M. L., Passarino, G. & Bellizzi, D. SIRT3 gene expression: a link between inherited mitochondrial DNA variants and oxidative stress. Gene 497, 323–329 (2012).
Dunbar, D. R., Moonie, P. A., Jacobs, H. T. & Holt, I. J. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc. Natl Acad. Sci. USA 92, 6562–6566 (1995).
Leslie, S. et al. The fine-scale genetic structure of the British population. Nature 519, 309–314 (2015).
Wei, W. et al. Germline selection shapes human mitochondrial DNA diversity. Science 364, eaau6520 (2019).
Latorre-Pellicer, A. et al. Regulation of mother-to-offspring transmission of mtDNA heteroplasmy. Cell Metab. 30, 1120–1130 (2019).
Latorre-Pellicer, A. et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature 535, 561–565 (2016).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Wain, L. V. et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir. Med. 3, 769–781 (2015).
Surendran, P. et al. Discovery of rare variants associated with blood pressure regulation through meta-analysis of 1.3 million individuals. Nat. Genet. 52, 1314–1332 (2020).
Weissensteiner, H. et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–W63 (2016).
van Oven, M. & Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30, E386–E394 (2009).
Calabrese, C. et al. MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics 30, 3115–3117 (2014).
Landrum, M. J. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
Chalkia, D. et al. Association between mitochondrial DNA haplogroup variation and autism spectrum disorders. JAMA Psychiatry 74, 1161–1168 (2017).
Hudson, G. et al. Two-stage association study and meta-analysis of mitochondrial DNA variants in Parkinson disease. Neurology 80, 2042–2048 (2013).
Kraja, A. T. et al. Associations of mitochondrial and nuclear mitochondrial variants and genes with seven metabolic traits. Am. J. Hum. Genet. 104, 112–138 (2019).
Zhan, X., Hu, Y., Li, B., Abecasis, G. R. & Liu, D. J. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 32, 1423–1426 (2016).
Ma, C., Blackwell, T., Boehnke, M. & Scott, L. J., GoT2D investigators. Recommended joint and meta-analysis strategies for case–control association testing of single low-count variants. Genet. Epidemiol. 37, 539–550 (2013).
Meyer, J. N., Hartman, J. H. & Mello, D. F. Mitochondrial toxicity. Toxicol. Sci. 162, 15–23 (2018).
Vial, G., Detaille, D. & Guigas, B. Role of mitochondria in the mechanism(s) of action of metformin. Front Endocrinol. 10, 294 (2019).
Astle, W. J. et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167, 1415–1429 (2016).
Sinnott-Armstrong, N. et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 53, 185–194 (2021).
Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016).
Benner, C. et al. Prospects of fine-mapping trait-associated genomic regions by using summary statistics from genome-wide association studies. Am. J. Hum. Genet. 101, 539–551 (2017).
Feng, S., Liu, D., Zhan, X., Wing, M. K. & Abecasis, G. R. RAREMETAL: fast and powerful meta-analysis for rare variants. Bioinformatics 30, 2828–2829 (2014).
Acknowledgements
We are grateful to: G. Hudson and H. Griffin for discussions and the preliminary exploratory work that proceeded this study; P. Surendran and T. Jiang for assistance with the genotype calling scripts; and W. Astle from the University of Cambridge for providing blood cell trait phenotypes and summary statistics. The BHF Cardiovascular Epidemiology Unit is supported by the UK Medical Research Council (MR/L003120/1), British Heart Foundation (RG/13/13/30194 and RG/18/13/33946) and the NIHR Cambridge Biomedical Research Center (BRC-1215-20014) and Health Data Research UK (which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome). P.C. is a Wellcome Trust Principal Research Fellow (212219/Z/18/Z) and a UK NIHR senior investigator, who receives support from the Medical Research Council Mitochondrial Biology Unit (MC_UU_00015/9), the Medical Research Council International Center for Genomic Medicine in Neuromuscular Disease, the Evelyn Trust and the NIHR Cambridge BRC (BRC-1215-20014) [*]. J.H. is funded by the British Heart Foundation (RG/13/13/30194) and the NIHR Cambridge BRC (BRC-1215-20014) [*]. E.Y.-D. was funded by the Isaac Newton Trust/Wellcome Trust ISSF/University of Cambridge Joint Research Grants Scheme. A.G.-D. is funded by the NIHR Cambridge BRC (146281). This research was conducted using the UKBB Resource under application numbers 20480, 7439 and 18794. [*]The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Author information
Authors and Affiliations
Contributions
E.Y.-D. and C.C. performed analyses and drafted the manuscript. K.S. and A.G.-D. selected binary traits for analysis and filtering. W.W. performed GenBank data retrieval and initial QC. S.K. performed initial QC of UKBB data. P.F.C. and J.M.M.H. drafted the manuscript and supervised the work. All authors approved the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
J.M.M.H. and E.Y.D. became full-time employees of Novo Nordisk during the drafting of the manuscript. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Valerio Carelli and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
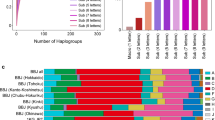
Extended Data Fig. 1 Distribution of mitochondrial sub-haplogroups across Great Britain.
The European unrelated individuals with birth coordinates (N = 327,665) were clustered based on the first 10 nucPCs, resulting in eight nuclear clusters. The map of Great Britain is colored according to the five regions identified by the most common clusters or combination of clusters in each region: (1) Scotland; (2) North of England (North East and West); (3) North of England (Yorkshire and the Humber, North West of England); (4) South of England (Midlands, London, South East and West of England); (5) Wales. No data were available for Northern Ireland. The stacked bar charts represent the frequency of unrelated individuals in each mitochondrial sub-haplogroups, in the five regions identified by the most common nuclear clusters or combination of nuclear clusters. The star indicates an over-representation (likelihood ratio test, two-sided P < 5 × 10−5) of J1b sub-haplogroup in Scotland compared to the Midlands, London, South East and West region.
Extended Data Fig. 2 Relationship between population structure in the nuclear and mitochondrial genomes.
The figure shows (a) circular Manhattan plots of the association between the first 10 nucPCs and mtSNVs. For each mtSNV, the association was tested using a linear regression model: Y~ β1 x X1 + β2 x X2 + β3 x X3 + β4 x X4 + β5 x X5 where Y is a vector containing the values of a nucPC, X1 is a vector of mtSNV dosages and X2-X5 are vectors containing covariate values (age, age squared, sex, and array) and β1-5 represent the effect of each variable on the mean of Y. Wald test two-sided P-values are presented. The nucPCs are ordered from PC1 to PC10 from outside to in and black dots represent (Wald test, two-sided) P < 5 × 10−5; (b) 3D plots of the first three mtPCs; and (c) the relationship between the first three nuclear principal components (nucPCs, nucPC1 - left, nucPC2 - middle, nucPC3 - right) and the first two mitochondrial principal components (mtPCs). The latter were calculated using mtSNVs with MAF > 0.01 and R2 < 0.2. The mtPCs in (a) and (b) were calculated using the following sets of genotyped mtSNV: (from left to right) all mtSNVs; mtSNVs with MAF > 0.01 only; and mtSNVs with MAF > 0.01 after LD-pruning at R2 < 0.2. N = the number of mtSNVs included in a given analysis. In (b) and (c) individuals are coloured according to macro-haplogroup carrier status.
Extended Data Fig. 3 Principal components analysis of the European set of UK Biobank participants in comparison to European participants in GenBank, 1000 genomes and WTCCC.
Plots of the first three mitochondrial principal components (mtPCs) for individuals in: (a) the European set of UK Biobank (N = 358,916), (b) GenBank reference set used for imputation (N = 6,593), (c) 1000 Genomes individuals (N = 498) and (d) WTCCC controls (N = 747). For each of the three data sets, plots on the left-hand side show mtPCs calculated using pruned SNVs (R2 < 0.2 for UK Biobank and R2 < 0.1 for GenBank, 1000 Genomes and WTCCC) while the plots on the right were generated without LD-pruning. Individuals are colored according to macro-haplogroup carrier status. mtPCs were calculated using genotyped SNVs (MAF > 0.01).
Supplementary information
Supplementary Information
Supplementary Note and Figs. 1–11
Rights and permissions
About this article
Cite this article
Yonova-Doing, E., Calabrese, C., Gomez-Duran, A. et al. An atlas of mitochondrial DNA genotype–phenotype associations in the UK Biobank. Nat Genet 53, 982–993 (2021). https://doi.org/10.1038/s41588-021-00868-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00868-1
This article is cited by
-
Cytoplasmic genome contributions to domestication and improvement of modern maize
BMC Biology (2024)
-
CD4+ T cell mitochondrial genotype in Multiple Sclerosis: a cross-sectional and longitudinal analysis
Scientific Reports (2024)
-
Mitochondrial genetic variation and risk of chronic kidney disease and acute kidney injury in UK Biobank participants
Human Genetics (2024)
-
Mitochondria-wide association study observed significant interactions of mitochondrial respiratory and the inflammatory in the development of anxiety and depression
Translational Psychiatry (2023)
-
The contributions of mitochondrial and nuclear mitochondrial genetic variation to neuroticism
Nature Communications (2023)