Abstract
Inactivating mutations in LKB1/STK11 are present in roughly 20% of nonsmall cell lung cancers (NSCLC) and portend poor response to anti-PD-1 immunotherapy. Unexpectedly, we found that LKB1 deficiency correlated with elevated tumor mutational burden (TMB) in NSCLCs from nonsmokers and genetically engineered mouse models, despite the frequent association between high-TMB and anti-PD-1 treatment efficacy. However, LKB1 deficiency also suppressed antigen processing and presentation, which are associated with compromised immunoproteasome activity and increased autophagic flux. Immunoproteasome activity and antigen presentation were restored by inhibiting autophagy through targeting the ATG1/ULK1 pathway. Accordingly, ULK1 inhibition synergized with PD-1 antibody blockade, provoking effector T-cell expansion and tumor regression in Lkb1-mutant tumor models. This study reveals an interplay between the immunoproteasome and autophagic catabolism in antigen processing and immune recognition, and proposes the therapeutic potential of dual ULK1 and PD-1 inhibition in LKB1-mutant NSCLC as a strategy to enhance antigen presentation and to promote antitumor immunity.
This is a preview of subscription content, access via your institution
Access options






Similar content being viewed by others
Data availability
All data generated and supporting the findings of this study are available within the paper. The RNA-seq data have been deposited in the Gene Expression Ominbus accession number GSE137244 and GSE137396. The whole-exome sequencing data have been deposited in the NCBI Sequence Read Archive accession number PRJNA564395. TCGA data used are publicly available at the Genomic Data Commons portal (https://portal.gdc.cancer.gov/). Source data are available for this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
Skoulidis, F. et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 8, 822–835 (2018).
Kadara, H. et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann. Oncol. 28, 75–82 (2017).
Rizvi, H. et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 36, 633–641 (2018).
Herter-Sprie, G. S. et al. Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer. JCI Insight 1, e87415 (2016).
Xu, C. et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell 25, 590–604 (2014).
Koyama, S. et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res. 76, 999–1008 (2016).
Deng, J. et al. CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov. 8, 216–233 (2018).
Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
McGranahan, N. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
McFadden, D. G. et al. Mutational landscape of EGFR-, MYC-, and Kras-driven genetically engineered mouse models of lung adenocarcinoma. Proc. Natl Acad. Sci. USA 113, E6409–E6417 (2016).
Cheng, D. T. et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Govindan, R. et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 150, 1121–1134 (2012).
Turajlic, S. et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 18, 1009–1021 (2017).
Mandal, R. et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 364, 485–491 (2019).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Davies, H. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 23, 517–525 (2017).
Helleday, T., Eshtad, S. & Nik-Zainal, S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15, 585–598 (2014).
Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999).
Bennardo, N., Cheng, A., Huang, N. & Stark, J. M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008).
Jasin, M. & Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5, a012740 (2013).
Panier, S. & Durocher, D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat. Rev. Mol. Cell Biol. 14, 661–672 (2013).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Nolan, E. et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aal4922 (2017).
Hisamatsu, H. et al. Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J. Exp. Med. 183, 1807–1816 (1996).
Boehm, U., Klamp, T., Groot, M. & Howard, J. C. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15, 749–795 (1997).
Seliger, B. et al. IFN-gamma-mediated coordinated transcriptional regulation of the human TAP-1 and LMP-2 genes in human renal cell carcinoma. Clin. Cancer Res. 3, 573–578 (1997).
Peaper, D. R., Wearsch, P. A. & Cresswell, P. Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. EMBO J. 24, 3613–3623 (2005).
Dikic, I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 (2017).
Poillet-Perez, L. et al. Autophagy maintains tumour growth through circulating arginine. Nature 563, 569–573 (2018).
Yang, A. et al. Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 8, 276–287 (2018).
Bhatt, V. et al. Autophagy modulates lipid metabolism to maintain metabolic flexibility for Lkb1-deficient Kras-driven lung tumorigenesis. Genes Dev. 33, 150–165 (2019).
Kim, H. S. et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell 155, 552–566 (2013).
Skoulidis, F. et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 5, 860–877 (2015).
Osorio, F., Lambrecht, B. N. & Janssens, S. Antigen presentation unfolded: identifying convergence points between the UPR and antigen presentation pathways. Curr. Opin. Immunol. 52, 100–107 (2018).
Shintani, T. & Klionsky, D. J. Autophagy in health and disease: a double-edged sword. Science 306, 990–995 (2004).
Petherick, K. J. et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 290, 28726 (2015).
Kimura, S., Noda, T. & Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007).
Kitajima, S. et al. Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discov. 9, 34–45 (2019).
Mazzucchelli, R. & Durum, S. K. Interleukin-7 receptor expression: intelligent design. Nat. Rev. Immunol. 7, 144–154 (2007).
Shackelford, D. B. & Shaw, R. J. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575 (2009).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Blokzijl, F., Janssen, R., van Boxtel, R. & Cuppen, E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 10, 33 (2018).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Bullard, J. H., Purdom, E., Hansen, K. D. & Dudoit, S. Evaluation of statistical methods for normalization and differential expression in mRNA-seq experiments. BMC Bioinf. 11, 94 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Reich, M. et al. GenePattern 2.0. Nat. Genet. 38, 500–501 (2006).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Chen, J., Bardes, E. E., Aronow, B. J. & Jegga, A. G. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 (2009).
Ji, H. et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 448, 807–810 (2007).
Yamamoto, K. et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581, 100–105 (2020).
Stark, J. M., Pierce, A. J., Oh, J., Pastink, A. & Jasin, M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 24, 9305–9316 (2004).
Acknowledgements
This work was supported by grants R01CA166480 (K.-K.W.), U01CA233084 (K.-K.W.), R01CA219670 (N.B., K.-K.W.), R01CA076584 (M.P.), SPORE P50CA058223 (C.M.P.), P50CA101942 (G.J.F.) and a fellowship from the T32 CA009161 (Levy) grant to A.M. We thank New York University (NYU) Langone Genome Technology Center for facilitating RNA-seq and whole-exome sequencing experiments. We thank the NYU Langone Division of Comparative Medicine staff for their support in the animal studies. We thank NYU Langone Health DART Microscopy Laboratory A. Liang, C. Petzold and K. Dancel-Manning for their assistance with TEM work. This core laboratory is partially funded by NYU Cancer Center Support grant no. NIH/NCI P30CA016087. M.P. is an Investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
J.D. and K.-K.W. conceived and designed the experiment. J.D. did most of the experiments described in this paper. A.T. and J.D. designed and performed the pathway, gene expression and GSEA analysis. I.D. and J.D. designed and performed mutational load and mutational signature analysis. J.T.P. performed specimen TMB score analysis on patients with NSCLC. J.D., H.S. and C.T. performed HR and microhomology-mediated end joining analysis. A.M. performed chromatin binding assay, immunoprecipitation and DSB foci analysis. M.B. and D.H.P. performed western blot for the antigen presentation pathway. M.B. and D.H.P. generated the Ulk1 shRNA and Atg cell lines. F.L. and H.Hu. generated the KL cell lines. F.L., Y.P. and H.D. generated and characterized KLP cell lines. J.D., H.D., D.H.P. and H.Han performed the immunoproteasome activity assay. J.D., T.C., E.P., V.P., C.T., S.L. and H.Hu. performed animal experiments, treatment studies and MRI imaging and analysis. J.D. and E.P. performed immune analysis of the animal models. T.C. performed the cell growth assay. J.L., J.D. and S.M. performed autophagy flux analysis. J.L. performed electron microscopy experiments and analysis. J.D., B.J. and N.S.G. participated in the Ulk1 inhibitor experiment. J.D. and N.B. drafted the paper. J.D., K.-K.W., N.B., V.W., E.S.W., P.S.H., N.S.G., T.P., A.T., M.Pagano, E.R., J.G., G.J.F., C.M.R., J.V.H., C.M.P., I.A. and M.Philips. conceptually designed and edited the paper. K.-K.W. conceived, designed and supervised all the experiments. All authors reviewed and discussed the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: K.-K.W. is a founder and equity holder of G1 Therapeutics. K.-K.W. has sponsored Research Agreements with MedImmune, Takeda, TargImmune, Mirati, Merus, Alkermes and BMS. K.-K.W. has consulting and sponsored research agreements with AstraZeneca, Janssen, Pfizer, Novartis, Merck, Ono, Array. C.M.P is an equity stock holder and consultant, and Board of Director Member of BioClassifier LLC and GeneCentric Diagnostics. C.M.P is also listed an inventor on patent applications on the Breast PAM50 and Lung Cancer Subtyping assays. C.M.R. has consulted regarding cancer drug development with AbbVie, Amgen, Ascentage, Bicycle, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Loxo and PharmaMar, and serves on the SAB of Bridge Medicines and Harpoon Therapeutics. M.Pagano is a cofounder of Coho Therapeutics; has financial interests in Coho Therapeutics, CullGen Inc. and Kymera Therapeutics; is on the SAB of CullGen Inc. and Kymera Therapeutics, and is a consultant for Coho Therapeutics, CullGen Inc., Kymera Therapeutics and SEED Therapeutics. J.F.G. has served as a compensated consultant or received honoraria from Bristol-Myers Squibb, Genentech, Ariad/Takeda, Loxo/Lilly, Blueprint, Oncorus, Regeneron, EMD Serono, Gilead, AstraZeneca, Pfizer, Incyte, Novartis, Merck, Agios, Amgen and Array; research support from Novartis, Genentech/Roche and Ariad/Takeda; institutional research support from Bristol-Myers Squibb, Tesaro, Moderna, Blueprint, Jounce, Array Biopharma, Merck, Adaptimmune, Novartis and Alexo; and has an immediate family member who is an employee of Ironwood Pharmaceuticals. G.J.F. has patents/pending royalties on the PD-1/PD-L1 pathway from Roche, Merck MSD, Bristol-Myers-Squibb, Merck KGA, Boehringer-Ingelheim, AstraZeneca, Dako, Leica, Mayo Clinic and Novartis. G.J.F. has served on advisory boards for Roche, Bristol-Myers-Squibb, Xios, Origimed, Triursus, iTeos, NextPoint, IgM, Jubilant and GV20. G.J.F. has equity in NextPoint, Triursus, Xios, iTeos, IgM and GV20.
Additional information
Peer review information Nature Cancer thanks Andrew Thorburn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Mutational burden and signature analysis for LKB1 mutant tumors.
a, Comparison of tumor mutational burden (TMB) of nonsynonymous mutation using freshly isolate NSCLC GEMMs lung nodules from KrasG12DLkb1fl/fl (KL) or KrasG12DTrp53fl/fl (KP) from RNA sequencing. n = 5 lung nod each group. (unpaired t-test, two sided, FDR < 0.05) b, TMB analysis of cell lines derived from KrasG12DLkb1fl/flTrp53fl/fl (KLP) NSCLC GEMM nodules using RNA sequencing (Upper table) or whole exome sequencing (WES) (lower table) for each cell line. c, Validation of KP, KL, KLP lines used. Upper panel, PCR result of indicated primers for each cell line. Lower panel, western blot result of LKB1, TRP53 and P16 protein levels of each cell lines. Blots are cropped and that uncropped images can be found in Source Data. Data represents one of three independent experiments. d, Representative IHC images of KL allograft lung tumors showing adenocarcinoma (left panels) or squamous tumor (right panels). Data represents one of three independent experiments. Scale bar, 100μm. e, Co-mutational analysis of NSCLC patients analyzed in Fig. 1b, including KRAS, STK11 and TP53 as total patient number and percentage within each group. f, Top suppressed biological process pathways from Kras/Lkb1 comparing with Kras/Trp53 mutant mouse cell lines. x-axis, -log2q value (Bonferroni) between the two groups. n = 5 cell lines each group.
Extended Data Fig. 2 Mutational signature analysis and gene set enrichment analysis (GSEA) from KL and KP tumors.
a, Point nonsynonymous single-nucleotide variations (SNV) numbers from either KP or KL cells. KP n = 6, KL n = 5 cell lines each group. (mean±sd, unpaired t test, two tailed). b, Percentage of Indels and SNVs in KP and KL cells KP n = 6, KL n = 5 cell lines each group. (unpaired t test, two sided. P = 0.0032). c, Percentage of each COSMIC mutational signature detected in the cell lines examined. Each column represents one individual cell line. d, Fold change of the signatures shown in (a) and normalized to average levels of corresponding signature in KP. KL, KP n = 5 cell lines each group. (mean ± sd, multiple t test, FDR < 0.05). e, GSEA of DNA repair related pathways, including HR repair-replication independent DSB, non-homologous end joining, nucleotide excision repair and mismatch repair, and related autophagy pathway and unfolded protein response (UPR) pathway of KL and KP GEMM lung nodules. KL n = 5, KP n = 5 lung nodules each group. f, Gene set enrichment analysis (GSEA) of TCGA patients for autophagy pathway and UPR pathway. KL n = 19, KP n = 22 patients each group.
Extended Data Fig. 3 LKB1 mutant tumor double strand break repair.
a, HR and NHEJ levels in Kras/Lkb1/Trp53 mutant cell lines comparing with Kras/Trp53 cells. n = 3 cell cultures for each group. (mean±sd, two-sided t test, unpaired). b, NHEJ ratio changes in human NSCLC LKB1 mutant cell lines H23, A427 and H460 with empty vector (pBABE), LKB1 or KLB1-KD overexpression were determined by flow cytometry. n = 3 individual cell cultures for each group. (mean±sd, two-sided t test, unpaired). c, Quantification of Rad51 and pH2AX positive cells percentage in KLP cells. Result is combined from three independent experiments. KP UT, KP NCS and KLP UT n = 4, KLP NCS n = 3 individual cell cultures each group. Data shown one of two independent experiments. (mean ± sd, one-way ANOVA, multiple two-sided comparison, Tukey test). d, LKB1 forms complex with BRCA1 in response to DNA damage. Upper panel, immunoprecipitation assay showing LKB1 interacting proteins in response to neocarzinostatin (NCS) treatment induced DNA damage. Lower panel, western blot showing total level of input proteins from whole-cell extract (WCE). Data represents one of two independent experiments. Blots are cropped and uncropped images can be found in Source Data.
Extended Data Fig. 4 Immunoproteasome activity changes in LKB1 mutant tumors upon IFNγ stimulation.
a, Real-time PCR showing antigen presentation genes expressing in LKB1 mutant lines with LKB1 overexpression. Left, H460 human NSCLC cells. Right, mouse KL cells with or without LKB1 over-expression. n = 3 experiments each group. (mean ± sd, unpaired t test, two tailed). *P < 0.05, ***P < 0.001. b, Western blot showing immunoproteasome subunits PSMB9 and PSMB8 levels, TAP1, Tapasin and B2M expression (Upper panel) and conventional proteasome subunits PSMB5 and PSMB6 expression (lower panel) from KP or KL lung nodules. Samples are derived from the same experiment and blots were processed in parallel. Blots are cropped and uncropped images can be found in Source Data. c, KL Ulk1 shRNA cell lines (upper panel) and KP Ulk1 shRNA cell lines (lower panel) were stimulated with IFN γ (10 ng/ml) for 24 hrs before the measurement of immunoproteasome activities showed as relative fluorescent units (RFU) per min (Vmax). n = 3 cell cultures for each group. (mean ± sd, multiple two-sided unpaired t test, two-stage step-up method of Benjamini, Krieger and Yekutieli). d, Immunoproteasome cleavage activity corresponding to indicated substrate for two KLP cells (KLP T1 and KLP T2) after IFN𝛾 stimulation shown as Vmax fold change compared with KL vehicle control group. n = 3 cell cultures each group. (mean ± sd, two-sided unpaired multiple t test, two-stage step-up method of Benjamini, Krieger and Yekutieli). e, Immunoproteasome activity for KL (left) and H460 (right) with or without LKB1 over-expression. n = 3 cell cultures each group. (mean ± sd, two-sided unpaired multiple t test, two-stage step-up method of Benjamini, Krieger and Yekutieli).
Extended Data Fig. 5 MHCI levels in KL and KP tumors.
a, MHCI levels from KP or KL allograft tumors as quantified as median fluorescent intensity (MFI) (left). Right, representative histograms depict MHCI level from mice with KP and KL tumors. n = 5 tumors each group. (mean ± sd, two-sided t test, unpaired) b, MHCI levels from KP or KL tumor cell lines stimulated with IFNγ as shown MFI levels. Left panel, representative histograms. Right panel, quantification of MHC I expression levels in KP and KL after IFNγ (100 ng/ml) stimulation for 18 hrs. n = 3 cell cultures each group. Shown representative result of three independent experiments (mean ± sd, multiple t test, FDR < 0.05). c, MHC I subunit H-2K and H-2D levels change in response to ULK1 inhibitor MRT68921 in mouse KL line in vitro 48hrs after the treatment. Data representative of two independent experiments.
Extended Data Fig. 6 Targeting autophagy pathway in LKB1 mutant NSCLC tumors.
a, Representative immunofluorescent (IF) image of autophagy flux in KL and KL-LKB1 tumor cells in response to MRT using GFP-mCherry-LC3B reporter. Red, GFP-mCherry+ LC3 puncta; yellow, GFP + mCherry+ puncta. scale bar, 15 𝜇m. Data representative of 2 independent experiments. b, Quantification of autophagic flux by GFP-RFP-LC3 reporter in KL tumors treated with MRT as left using flow cytometry analysis. Upper panel, gating strategy. Lower panel, quantification of autophagic flux shown as RFP:GFP ratios. n = 3 cell cultures each group. Data shown represents one of three independent experiments (mean ± sd, two-tailed t test for high group, unpaired). c, Quantification of autophagic flux ratio (RFP:GFP) in KL cells with GFP-RFP-LC3 reporter transduced with Ulk1 shRNA. n = 3 cell cultures each group. Data shown one of two independent experiments. (mean ± sd, two-tailed t test for high group, unpaired). Samples were compared with NT shRNA cells for high group. d. western blot showing ULK1 protein levels in KL (left) and KP (right) stable cell lines with Ulk1 shRNA. Data represents one of two independent experiments. e, Western blot of ULK inhibitor MRT68921 in human LKB1 isogenic lines for TBK1/STING pathway changes of H460 (left) and H23 (middle) and H1792 (right). Data represents one of two independent experiments. f,g,h, Immunoproteasome activity for KL stable cell lines with Atg7 or Atg13 for ANW (f), KQL (g) or PAL (h) substrate. n = 3 independent experimental samples for each group. (data presented are shown as mean±sd, two-sided one-way ANOVA, Tukey test, statistics presented on top of each column is compared with vehicle group, and pairwise comparisons between groups after 100 ng/ml IFNγ treatment shown on top of columns). Data shown one of two independent experiments. i, Western blot of KL stable cell lines generated with either Atg7 shRNA (left panel) or Atg13 shRNA (right panel). Shown one of two independent experiments. Blots in panels d, e and i are cropped and uncropped images can be found in Source Data.
Extended Data Fig. 7 Targeting autophagy in LKB1 mutant NSCLC tumors in vivo.
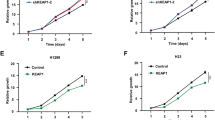
a, validation of KL cells transduced with autophagy flux reporter GFP–LC3-RFP (GLR). Generated KL GLR cells were starved in EBSS buffer for 24 hrs before FACS analysis for RFP/GFP signals. b, Representative FACS analysis of GFP/RFP ratio of KL tumors after indicated treatments. Data representative of 2 independent experiments. c, Western blot shows conventional proteasome subunits PSMB5 and PSMB6 expression levels from KL lung nodules after MRT + PD1 treatment. Each lane represents one individual mouse tumor nodule. d, Toxicity of MRT68921 shown as mouse body weight change percentage after the drug treatment. Each line represents one mouse. n = 9. e, Tumor volume changes after 2-week treatment of KL GEMM tumors with MRT + PD1. Veh n = 9, PD1 n = 14, MRT n = 8, MRT + PD1, n = 14 tumors each group. (mean ± sd, one-way ANOVA). f, Tumor volume change of KP allograft tumor 1 week after indicated treatment. Veh, MRT, MRT + PD1 n = 10; PD1 n = 8 tumors each group. (mean ± sd, one-way ANOVA).
Extended Data Fig. 8 Immune infiltrates analysis in KL tumors after ULK1 inhibitor treatment.
a, Gating strategy used for immune analysis. b, FACS analysis of CD4 + (left) and CD8 + (right) T lymphocytes among total tumor infiltrating CD45 + leukocytes in KL tumor after the treatment of ULK1 inhibitor MRT68921 (MRT). Veh n = 12, MRT n = 5 tumors each group. (mean ± sd, unpaired t test, two tailed). c, CD8/Treg ratio of total tumor infiltrating leukocytes (TILs) after the indicated drugs treatment. veh n = 12, MRT n = 5, MRT + PD1 n = 12 tumors each group. (mean ± sd, unpaired t test, two tailed). d, CD69, CCR7 and 2B4 levels within CD8 + T cells after MRT68921 (MRT) and anti-PD1 treatment. Left panel, veh n = 12, MRT + PD1 n = 10. Middle and right panel, veh n = 8, MRT + PD1 n = 10 tumors each group. (mean ± sd, unpaired t test, two tailed.).
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Deng, J., Thennavan, A., Dolgalev, I. et al. ULK1 inhibition overcomes compromised antigen presentation and restores antitumor immunity in LKB1-mutant lung cancer. Nat Cancer 2, 503–514 (2021). https://doi.org/10.1038/s43018-021-00208-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-021-00208-6
This article is cited by
-
Carnosine regulation of intracellular pH homeostasis promotes lysosome-dependent tumor immunoevasion
Nature Immunology (2024)
-
Lysosomes as coordinators of cellular catabolism, metabolic signalling and organ physiology
Nature Reviews Molecular Cell Biology (2024)
-
Exploiting autophagy balance in T and NK cells as a new strategy to implement adoptive cell therapies
Molecular Cancer (2023)
-
Impact of context-dependent autophagy states on tumor progression
Nature Cancer (2023)
-
At the crossroads of immunotherapy for oncogene-addicted subsets of NSCLC
Nature Reviews Clinical Oncology (2023)