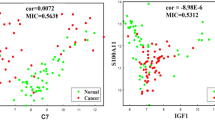
Abstract
Gene co-expression networks (GCN) present undirected relations between genes to understand molecular structures behind the diseases, including cancer. The utilization of various biological datasets and gene network inference (GNI) algorithms can reveal meaningful gene–gene interactions of GCNs. This study applies three GNI algorithms on mRNA gene expression, RNA-Seq, and miRNA–target genes datasets to infer GCNs of breast and prostate cancers. To evaluate the performance of the GCNs, we utilize overlap analysis via literature data, topological assessment, and Gene Ontology-based biological assessment. The results emphasize how the selection of biological datasets and GNI algorithms affect the performance results on different evaluation criteria. GCNs on microarray gene expression data slightly outperform in overlap analysis. Also, GCNs on RNA-Seq and gene expression datasets follow scale-free topology. The biological assessment results are close to each other on all biological datasets. C3NET algorithm-based GCNs did not contain any biological assessment modules; therefore, it is not optimal for biological assessment. GNI algorithms' selection did not change the overlap analysis and topological assessment results. Our primary objective is to compare the performance results of biological datasets and GNI algorithms based on different evaluation criteria. For this purpose, we developed the GNIAP R package that enables users to select different GNI algorithms to infer GCNs. The GNIAP R package also provides literature-based overlap analysis, and topological and biological analyses on GCNs. Users can access the GNIAP R package via https://github.com/ozgurcingiz/GNIAP.
Graphic abstract








Similar content being viewed by others
References
Ruggero D (2013) Translational control in cancer etiology. Cold Spring Harb Perspect Biol 5(2):a012336. https://doi.org/10.1101/cshperspect.a012336
Rao MS, Van Vleet TR, Ciurlionis R, Buck WR, Mittelstadt SW, Blomme EAG, Liguori MJ (2019) Comparison of RNA-Seq And microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front Genet 9:636. https://doi.org/10.3389/fgene.2018.00636
Xu X, Zhang Y, Williams J, Antoniou E, McCombie WR, Wu S, Zhu W, Davidson NO, Denoya P, Li E (2013) Parallel comparison of Illumina RNA-Seq and Affymetrix microarray platforms on transcriptomic profiles generated from 5-aza-deoxy-cytidine treated HT-29 colon cancer cells and simulated datasets. BMC Bioinform 14(Suppl 9):S1. https://doi.org/10.1186/1471-2105-14-S9-S1
Hung JH, Weng Z (2017) Analysis of microarray and RNA-seq expression profiling data. Cold Spring Harb Protoc. https://doi.org/10.1101/pdb.top093104
Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A (2016) A survey of best practices for RNA-seq data analysis. Genome Biol 17:13. https://doi.org/10.1186/s13059-016-0881-8
Mathur R, Rotroff D, Ma J, Shojaie A, Motsinger-Reif A (2018) Gene set analysis methods: a systematic comparison. BioData Min 11:8. https://doi.org/10.1186/s13040-018-0166-8
Li H, Wang N, Gong P, Perkins EJ, Zhang C (2011) Learning the structure of gene regulatory networks from time series gene expression data. BMC Genomics 12(Suppl 5):S13. https://doi.org/10.1186/1471-2164-12-S5-S13
Liu F, Zhang SW, Guo WF, Wei ZG, Chen L (2016) Inference of gene regulatory network based on local Bayesian networks. PLoS Comput Biol 12(8):e1005024. https://doi.org/10.1371/journal.pcbi.1005024
Singh N, Vidyasagar M (2015) bLARS: an algorithm to infer gene regulatory networks. IEEE/ACM Trans Comput Biol Bioinform 13(2):301–314. https://doi.org/10.1109/TCBB.2015.2450740
Ozsolak F, Milos PM (2011) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12(2):87–98. https://doi.org/10.1038/nrg2934
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578. https://doi.org/10.1038/nprot.2012.016
Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T (2017) Transcriptomics technologies. PLoS Comput Biol 13(5):e1005457. https://doi.org/10.1371/journal.pcbi.1005457
Ballouz S, Verleyen W, Gillis J (2015) Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinform 31(13):2123–2130. https://doi.org/10.1093/bioinformatics/btv118
de Matos SR, Dalleau S, Williamson KE, Emmert-Streib F (2015) Urothelial cancer gene regulatory networks inferred from large-scale RNAseq, Bead and Oligo gene expression data. BMC Syst Biol 9:21. https://doi.org/10.1186/s12918-015-0165-z
Iancu OD, Kawane S, Bottomly D, Searles R, Hitzemann R, McWeeney S (2012) Utilizing RNA-Seq data for de novo co-expression network inference. Bioinform 28(12):1592–1597. https://doi.org/10.1093/bioinformatics/bts245
Giorgi FM, Del Fabbro C, Licausi F (2013) Comparative study of RNA-seq- and microarray-derived co-expression networks in Arabidopsis thaliana. Bioinform 29(6):717–724. https://doi.org/10.1093/bioinformatics/btt053
Chang YM, Lin HH, Liu WY, Yu CP, Chen HJ, Wartini PP, Kao YY, Wu YH, Lin JJ, Lu MJ, Tu SL, Wu SH, Shiu SH, Ku MSB, Li WH (2019) Comparative transcriptomics method to infer gene co-expression networks and its applications to maize and rice leaf transcriptomes. Proc Natl Acad Sci U S A 116(8):3091–3099. https://doi.org/10.1073/pnas.1817621116
Gennarino VA, D’Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, Mutarelli M, Belcastro V, Ballabio A, Verde P, Sardiello M, Banfi S (2012) Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res 22(6):1163–1172. https://doi.org/10.1101/gr.130435.111
Cingiz MÖ, Biricik G, Diri B (2017) ARNetMiT R Package: association rules based gene co-expression networks of miRNA targets. Cell Mol Biol (Noisy-le-grand) 63(3):18–25. https://doi.org/10.14715/cmb/2017.63.3.4
Casey T, Bond J, Tighe S, Hunter T, Lintault L, Patel O, Eneman J, Crocker A, White J, Tessitore J, Stanley M, Harlow S, Weaver D, Muss H, Plaut K (2009) Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast Cancer Res Treat 114(1):47–62. https://doi.org/10.1007/s10549-008-9982-8
Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, Stephens RM, Caporaso NE, Loffredo CA, Ambs S (2008) Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res 68(3):927–936. https://doi.org/10.1158/0008-5472.CAN-07-2608
Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, Schellen P, Verschueren H, Post E, Koster J, Ylstra B, Ameziane N, Dorsman J, Smit EF, Verheul HM, Noske DP, Reijneveld JC, Nilsson RJA, Tannous BA, Wesseling P, Wurdinger T (2015) RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 28(5):666–676. https://doi.org/10.1016/j.ccell.2015.09.018
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium (2020) Pan-cancer analysis of whole genomes. Nature 578(7793):82–93. https://doi.org/10.1038/s41586-020-1969-6
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, Chien CH, Wu MC, Huang CY, Tsou AP, Huang HD (2011) miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 39(Database issue):D163–D169. https://doi.org/10.1093/nar/gkq1107
Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y (2008) miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 37(Database issue):D98–D104. https://doi.org/10.1093/nar/gkn714
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy–analysis of Affymetrix GeneChip data at the probe level. Bioinform 20(3):307–315. https://doi.org/10.1093/bioinformatics/btg405
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinform 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A (2006) ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform 7(Suppl 1):S7. https://doi.org/10.1186/1471-2105-7-S1-S7
Altay G, Emmert-Streib F (2010) Inferring the conservative causal core of gene regulatory networks. BMC Syst Biol 4:132. https://doi.org/10.1186/1752-0509-4-132
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 9:559. https://doi.org/10.1186/1471-2105-9-559
Meyer PE, Lafitte F, Bontempi G (2008) minet: A R/Bioconductor package for inferring large transcriptional networks using mutual information. BMC Bioinform 9:461. https://doi.org/10.1186/1471-2105-9-461
Niemira M, Collin F, Szalkowska A, Bielska A, Chwialkowska K, Reszec J, Niklinski J, Kwasniewski M, Kretowski A (2019) Molecular signature of subtypes of non-small-cell lung cancer by large-scale transcriptional profiling: identification of key modules and genes by weighted gene co-expression network analysis (WGCNA). Cancers 12(1):37. https://doi.org/10.3390/cancers12010037
Wang M, Wang L, Pu L, Li K, Feng T, Zheng P, Li S, Sun M, Yao Y, Jin L (2020) LncRNAs related key pathways and genes in ischemic stroke by weighted gene co-expression network analysis (WGCNA). Genomics 112(3):2302–2308. https://doi.org/10.1016/j.ygeno.2020.01.001
Dorantes-Gilardi R, García-Cortés D, Hernández-Lemus E, Espinal-Enríquez J (2020) Multilayer approach reveals organizational principles disrupted in breast cancer co-expression networks. Appl Netw Sci 5:47. https://doi.org/10.1007/s41109-020-00291-1
Nazanin H, Allahverdi A, Abdolmeki F (2020) The novel potential multidrug-resistance biomarkers for Pseudomonas aeruginosa lung infections using transcriptomics data analysis. Inform Med Unlocked 22:100509. https://doi.org/10.1016/j.imu.2020.100509
Altay G, Altay N, Neal D (2013) Global assessment of network inference algorithms based on available literature of gene/protein interactions. Turk J Biol 37:547–555. https://doi.org/10.3906/biy-1210-8
Chung F, Lu L (2002) Connected components in random graphs with given expected degree sequences. Ann Comb 6:125–145. https://doi.org/10.1007/PL00012580
Szczepińska T, Pawłowski K (2013) Genomic positions of co-expressed genes: echoes of chromosome organisation in gene expression data. BMC Res Notes 6:229. https://doi.org/10.1186/1756-0500-6-229
Serin EA, Nijveen H, Hilhorst HW, Ligterink W (2016) Learning from co-expression networks: possibilities and challenges. Front Plant Sci 7:444. https://doi.org/10.3389/fpls.2016.00444
Lim D, Lee SH, Kim NK, Cho YM, Chai HH, Seong HH, Kim H (2013) Gene co-expression analysis to characterize genes related to marbling trait in Hanwoo (Korean) Cattle. Asian-Australas J Anim Sci 26(1):19–29. https://doi.org/10.5713/ajas.2012.12375
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform 4:2. https://doi.org/10.1186/1471-2105-4-2
Wu XY, Xia XY (2015) ProNet: biological network construction, visualization and analyses. R package version 1.0.0. https://rdrr.io/cran/ProNet/ Accessed 08 April 2021
Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33(Database issue):D514–D517. https://doi.org/10.1093/nar/gki033
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44(W1):W90–W97. https://doi.org/10.1093/nar/gkw377
Reimand J, Arak T, Vilo J (2011) g:Profiler–a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res 39(Web Server issue):W307–W315. https://doi.org/10.1093/nar/gkr378
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cingiz, M.Ö., Biricik, G. & Diri, B. The Performance Comparison of Gene Co-expression Networks of Breast and Prostate Cancer using Different Selection Criteria. Interdiscip Sci Comput Life Sci 13, 500–510 (2021). https://doi.org/10.1007/s12539-021-00440-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-021-00440-9