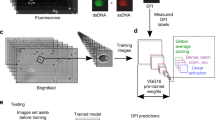
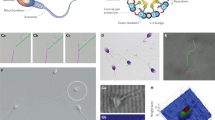
Abstract
Infertility rates and the number of couples seeking fertility care have increased worldwide over the past few decades. Over 2.5 million cycles of assisted reproductive technologies are being performed globally every year, but the success rate has remained at ~33%. Machine learning, an automated method of data analysis based on patterns and inference, is increasingly being deployed within the health-care sector to improve diagnostics and therapeutics. This technique is already aiding embryo selection in some fertility clinics, and has also been applied in research laboratories to improve sperm analysis and selection. Tremendous opportunities exist for machine learning to advance male fertility treatments. The fundamental challenge of sperm selection — selecting the most promising candidate from 108 gametes — presents a challenge that is uniquely well-suited to the high-throughput capabilities of machine learning algorithms paired with modern data processing capabilities.
Key points
-
Selection of sperm during intracytoplasmic sperm injection is currently not standardized as the WHO guidelines are interpreted subjectively by clinical experts.
-
Machine learning shows promise in improving intracytoplasmic sperm injection by guiding the clinicians to objectively select sperm.
-
Most reported machine learning methods for classification of the sperm head rely on data labelled by clinical experts, despite the variability that exists between them.
-
New studies on single sperm motility and DNA integrity can be used as unbiased quantitative measures for machine learning algorithm-based sperm selection.
-
Considerations of data quality, subjectivity in training data and representativeness of data are crucial to minimize bias.
-
Ethical questions, such as data privacy, control over the decision-making process and data variability, warrant further consideration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Inhorn, M. C. & Patrizio, P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum. Reprod. Update 21, 411–426 (2015).
Agarwal, A., Mulgund, A., Hamada, A. & Chyatte, M. R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 13, 37 (2015).
Hammoud, A. et al. Decreased sperm motility is associated with air pollution in Salt Lake City. Fertil. Steril. 93, 1875–1879 (2010).
Jurewicz, J., Hanke, W., Radwan, M. & Bonde, J. Environmental factors and semen quality. Int. J. Occup. Med. Environ. Health 22, 305–329 (2009).
Lafuente, R., García-Blàquez, N., Jacquemin, B. & Checa, M. A. Outdoor air pollution and sperm quality. Fertil. Steril. 106, 880–896 (2016).
Jensen, T. K. et al. High dietary intake of saturated fat is associated with reduced semen quality among 701 young Danish men from the general population. Am. J. Clin. Nutr. 97, 411–418 (2013).
Afeiche, M. et al. Dairy food intake in relation to semen quality and reproductive hormone levels among physically active young men. Hum. Reprod. 28, 2265–2275 (2013).
Hammoud, A. O. Obesity and male reproductive potential. J. Androl. 27, 619–626 (2006).
Du Plessis, S. S., Cabler, S., McAlister, D. A., Sabanegh, E. & Agarwal, A. The effect of obesity on sperm disorders and male infertility. Nat. Rev. Urol. 7, 153–161 (2010).
Mascarenhas, M. N., Flaxman, S. R., Boerma, T., Vanderpoel, S. & Stevens, G. A. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med. 9, e1001356 (2012).
Boivin, J., Bunting, L., Collins, J. A. & Nygren, K. G. International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum. Reprod. 22, 1506–1512 (2007).
Sear, R., Lawson, D. W., Kaplan, H. & Shenk, M. K. Understanding variation in human fertility: what can we learn from evolutionary demography? Phil. Trans. R. Soc. B 371, 20150144 (2016).
Fauser, B. C. Towards the global coverage of a unified registry of IVF outcomes. Reprod. Biomed. Online 38, 133–137 (2019).
Nosrati, R. et al. Microfluidics for sperm analysis and selection. Nat. Rev. Urol. 14, 707–730 (2017).
Wilkinson, J. et al. Reproductive medicine: still more ART than science? BJOG 126, 138–141 (2019).
Oseguera-López, I., Ruiz-Díaz, S., Ramos-Ibeas, P. & Pérez-Cerezales, S. Novel techniques of sperm selection for improving IVF and ICSI outcomes. Front. Cell Dev. Biol. 7, 298 (2019).
Swain, J. E. & Pool, T. B. ART failure: oocyte contributions to unsuccessful fertilization. Hum. Reprod. Update 14, 431–446 (2008).
Nasr-Esfahani, M. H., Deemeh, M. R. & Tavalaee, M. New era in sperm selection for ICSI. Int. J. Androl. 35, 475–484 (2012).
Bungum, M. & Oleszczuk, K. in A Clinician’s Guide to Sperm DNA and Chromatin Damage (eds Zini, A. & Agarwal, A.) 393–410 (Springer, 2018).
Claassens, O. E., Menkveld, R. & Harrison, K. L. Evaluation of three substitutes for Percoll in sperm isolation by density gradient centrifugation. Hum. Reprod. 13, 3139–3143 (1998).
Rappa, K. L. et al. Sperm processing for advanced reproductive technologies: where are we today? Biotechnol. Adv. 34, 578–587 (2015).
Younglai, E. V., Holt, D., Brown, P., Jurisicova, A. & Casper, R. F. Sperm swim-up techniques and DNA fragmentation. Hum. Reprod. 16, 1950–1953 (2001).
Jayaraman, V., Upadhya, D., Narayan, P. K. & Adiga, S. K. Sperm processing by swim-up and density gradient is effective in elimination of sperm with DNA damage. J. Assist. Reprod. Genet. 29, 557–563 (2012).
Yamanaka, M. et al. Combination of density gradient centrifugation and swim-up methods effectively decreases morphologically abnormal sperms. J. Reprod. Dev. 62, 599–606 (2016).
Repping, S., van Weert, J.-M., Mol, B. W., de Vries, J. W. & van der Veen, F. Use of the total motile sperm count to predict total fertilization failure in in vitro fertilization. Fertil. Steril. 78, 22–28 (2002).
Cooper, T. G. et al. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update 16, 231–245 (2010).
World Health Organization. WHO laboratory manual for the examination and processing of human semen (WHO, 2010).
Barroso, G. et al. Intra- and inter-laboratory variability in the assessment of sperm morphology by strict criteria: impact of semen preparation, staining techniques and manual versus computerized analysis. Hum. Reprod. 14, 2036–2040 (1999).
Dominguez, E. M., Moreno-Irusta, A., Guidobaldi, H. A., Tribulo, H. & Giojalas, L. C. Improved bovine in vitro embryo production with sexed and unsexed sperm selected by chemotaxis. Theriogenology 122, 1–8 (2018).
Bahat, A. et al. Thermotaxis of mammalian sperm cells: a potential navigation mechanism in the female genital tract. Nat. Med. 9, 149–150 (2003).
Amann, R. P. & Waberski, D. Computer-assisted sperm analysis (CASA): capabilities and potential developments. Theriogenology 81, 5–17 (2014).
Collins, J. A., Barnhart, K. T. & Schlegel, P. N. Do sperm DNA integrity tests predict pregnancy with in vitro fertilization? Fertil. Steril. 89, 823–831 (2008).
Cipolla, R, Battiato, S. & Farinella, G. M. (eds) Machine Learning for Computer Vision Vol. 411 (Springer, 2013).
Haines, N., Southward, M. W., Cheavens, J. S., Beauchaine, T. & Ahn, W.-Y. Using computer-vision and machine learning to automate facial coding of positive and negative affect intensity. PLoS ONE 14, e0211735 (2019).
Diehl, P. U. & Cook, M. Unsupervised learning of digit recognition using spike-timing-dependent plasticity. Front. Comput. Neurosci. 9, 99 (2015).
Beam, A. L. & Kohane, I. S. Big data and machine learning in health care. JAMA 319, 1317 (2018).
Sajda, P. Machine learning for detection and diagnosis of disease. Annu. Rev. Biomed. Eng. 8, 537–565 (2006).
Banaee, H., Ahmed, M. & Loutfi, A. Data mining for wearable sensors in health monitoring systems: a review of recent trends and challenges. Sensors 13, 17472–17500 (2013).
Goldenberg, S. L., Nir, G. & Salcudean, S. E. A new era: artificial intelligence and machine learning in prostate cancer. Nat. Rev. Urol. 16, 391–403 (2019).
Im, H. et al. Design and clinical validation of a point-of-care device for the diagnosis of lymphoma via contrast-enhanced microholography and machine learning. Nat. Biomed. Eng. 2, 666–674 (2018).
Lo, Y.-C., Rensi, S. E., Torng, W. & Altman, R. B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 23, 1538–1546 (2018).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Liu, Y. & Zhang, M. Neural network methods for natural language processing. Comput. Linguist. 44, 193–195 (2018).
Hirschberg, J. & Manning, C. D. Advances in natural language processing. Science 349, 261–266 (2015).
Wang, R. et al. Artificial intelligence in reproductive medicine. Reproduction 158, R139–R154 (2019).
Chu, K. Y. et al. Artificial intelligence in reproductive urology. Curr. Urol. Rep. 20, 52 (2019).
Thirumalaraju, P. et al. Deep learning-enabled blastocyst prediction system for cleavage stage embryo selection. Fertil. Steril. 111, e29 (2019).
Khosravi, P. et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit. Med. 2, 21 (2019).
Boulet, S. L. et al. Trends in use of and reproductive outcomes associated with intracytoplasmic sperm injection. JAMA 313, 255–263 (2015).
Dyer, S. et al. International committee for monitoring assisted reproductive technologies world report: assisted reproductive technology 2008, 2009 and 2010. Hum. Reprod. 31, 1588–1609 (2016).
Esteves, S. C., Roque, M., Bedoschi, G., Haahr, T. & Humaidan, P. Intracytoplasmic sperm injection for male infertility and consequences for offspring. Nat. Rev. Urol. 15, 535–562 (2018).
McCallum, C. et al. Deep learning-based selection of human sperm with high DNA integrity. Commun. Biol. 2, 250 (2019).
Menkveld, R., Holleboom, C. A. G. & Rhemrev, J. P. T. Measurement and significance of sperm morphology. Asian J. Androl. 13, 59–68 (2011).
Kruger, T. F. et al. New method of evaluating sperm morphology with predictive value for human in vitro fertilization. Urology 30, 248–251 (1987).
Gatimel, N., Moreau, J., Parinaud, J. & Léandri, R. D. Sperm morphology: assessment, pathophysiology, clinical relevance, and state of the art in 2017. Andrology 5, 845–862 (2017).
Brito, L. F. C. A multilaboratory study on the variability of bovine semen analysis. Theriogenology 85, 254–266 (2016).
Eustache, F. & Auger, J. Inter-individual variability in the morphological assessment of human sperm: effect of the level of experience and the use of standard methods. Hum. Reprod. 18, 1018–1022 (2003).
Singh, S., Sharma, S., Jain, M. & Chauhan, R. Importance of Papanicolaou Staining for Sperm Morphologic Analysis. Am. J. Clin. Pathol. 136, 247–251 (2011).
Schirren, C., Eckhardt, U., Jachczik, R. & Carstensen, C. A. Morphological differentiation of human spermatozoa with testsimplets® slides. Andrologia 9, 191–192 (2009).
Kruger, T. F. et al. A quick, reliable staining technique for human sperm morphology. Arch. Androl. 18, 275–277 (1987).
van der Horst, G. & Maree, L. SpermBlue®: A new universal stain for human and animal sperm which is also amenable to automated sperm morphology analysis. Biotech. Histochem. 84, 299–308 (2010).
Henkel, R. et al. Comparison of three staining methods for the morphological evaluation of human spermatozoa. Fertil. Steril. 89, 449–455 (2008).
Maree, L., du Plessis, S. S., Menkveld, R. & van der Horst, G. Morphometric dimensions of the human sperm head depend on the staining method used. Hum. Reprod. 25, 1369–1382 (2010).
Natali, I. et al. Scoring human sperm morphology using Testsimplets and Diff-Quik slides. Fertil. Steril. 99, 1227–1232.e2 (2013).
Czubaszek, M., Andraszek, K., Banaszewska, D. & Walczak-Jędrzejowska, R. The effect of the staining technique on morphological and morphometric parameters of boar sperm. PLoS ONE 14, e0214243 (2019).
Kruger, T. F. et al. A new computerized method of reading sperm morphology (strict criteria) is as efficient as technician reading. Fertil. Steril. 59, 202–209 (1993).
Kruger, T. F. et al. A prospective study on the predictive value of normal sperm morphology as evaluated by computer (IVOS**Hamilton Thome Research Version 2.1 Dimension Program, Beverly, Massachusetts.). Fertil. Steril. 66, 285–291 (1996).
Coetzee, K., de Villiers, A., Kruger, T. F. & Lombard, C. J. Clinical value of using an automated sperm morphology analyzer (IVOS). Fertil. Steril. 71, 222–225 (1999).
Coetzee, K., Kruger, T. F. & Lombard, C. J. Repeatability and variance analysis on multiple computer-assisted (IVOS*) sperm morphology readings. Andrologia 31, 163–168 (1999).
Menkveld, R. et al. Effects of different staining and washing procedures on the results of human sperm morphology evaluation by manual and computerised methods. Andrologia 29, 1–7 (2009).
Coetzee, K., Kruger, T. F., Vandendael, A., Villiers, A. & Lombard, C. J. Comparison of two staining and evaluation methods used for computerized human sperm morphology evaluations. Andrologia 29, 133–135 (2009).
Tseng, K.-K. et al. Computer-assisted system with multiple feature fused support vector machine for sperm morphology diagnosis. Biomed. Res. Int. 2013, 1–13 (2013).
Auger, J. et al. Sperm morphological defects related to environment, lifestyle and medical history of 1001 male partners of pregnant women from four European cities. Hum. Reprod. 16, 2710–2717 (2001).
Shaker, F., Monadjemi, S. A., Alirezaie, J. & Naghsh-Nilchi, A. R. A dictionary learning approach for human sperm heads classification. Comput. Biol. Med. 91, 181–190 (2017).
Chang, V., Garcia, A., Hitschfeld, N. & Härtel, S. Gold-standard for computer-assisted morphological sperm analysis. Comput. Biol. Med. 83, 143–150 (2017).
Chang, V., Heutte, L., Petitjean, C., Härtel, S. & Hitschfeld, N. Automatic classification of human sperm head morphology. Comput. Biol. Med. 84, 205–216 (2017).
Van Raemdonck, L. E. M. et al. An algorithm for morphological classification of motile human sperm (IEEE, 2015).
Javadi, S. & Mirroshandel, S. A. A novel deep learning method for automatic assessment of human sperm images. Comput. Biol. Med. 109, 182–194 (2019).
Riordon, J., McCallum, C. & Sinton, D. Deep learning for the classification of human sperm. Comput. Biol. Med. 111, 103342 (2019).
Ghasemian, F., Mirroshandel, S. A., Monji-Azad, S., Azarnia, M. & Zahiri, Z. An efficient method for automatic morphological abnormality detection from human sperm images. Comput. Methods Prog. Biomed. 122, 409–420 (2015).
Kanakasabapathy, M. K. et al. An automated smartphone-based diagnostic assay for point-of-care semen analysis. Sci. Transl Med. 9, 1–14 (2017).
Thirumalaraju, P. et al. Human sperm morphology analysis using smartphone microscopy and deep learning. Fertil. Steril. 112, e41 (2019).
Suarez, S. S. & Pacey, A. A. Sperm transport in the female reproductive tract. Hum. Reprod. Update 12, 23–37 (2006).
Eisenbach, M. & Giojalas, L. C. Sperm guidance in mammals — an unpaved road to the egg. Nat. Rev. Mol. Cell Biol. 7, 276–285 (2006).
Grunewald, S. & Paasch, U. in Male Infertility (eds Parekattil, S. & Agarwal, A.) 423–430 (Springer, 2012).
Sakkas, D., Ramalingam, M., Garrido, N. & Barratt, C. L. R. Sperm selection in natural conception: what can we learn from Mother Nature to improve assisted reproduction outcomes? Hum. Reprod. 21, 711–726 (2018).
Zini, A., Bielecki, R., Phang, D. & Zenzes, M. T. Correlations between two markers of sperm DNA integrity, DNA denaturation and DNA fragmentation, in fertile and infertile men. Fertil. Steril. 75, 674–677 (2001).
Wang, Y. et al. Prediction of DNA integrity from morphological parameters using a single-sperm DNA fragmentation index assay. Adv. Sci. 6, 1900712 (2019).
Evenson, D. P. The sperm chromatin structure assay (SCSA®) and other sperm DNA fragmentation tests for evaluation of sperm nuclear DNA integrity as related to fertility. Anim. Reprod. Sci. 169, 56–75 (2016).
Evenson, D. P., Larson, K. L. & Jost, L. K. Sperm chromatin structure assay: its clinical use for detecting sperm DNA fragmentation in male infertility and comparisons with other techniques. J. Androl. 23, 25–43 (2002).
Bungum, M. et al. The predictive value of sperm chromatin structure assay (SCSA) parameters for the outcome of intrauterine insemination, IVF and ICSI. Hum. Reprod. 19, 1401–1408 (2004).
Carell, D. & Kenneth, K. I. Spermatogenesis: Methods and Protocols (Humana, 2013).
Fernández, J. L. et al. The sperm chromatin dispersion test: a simple method for the determination of sperm DNA fragmentation. J. Androl. 24, 59–66 (2003).
Muriel, L. et al. Value of the sperm deoxyribonucleic acid fragmentation level, as measured by the sperm chromatin dispersion test, in the outcome of in vitro fertilization and intracytoplasmic sperm injection. Fertil. Steril. 85, 371–383 (2006).
Sun, T. C. et al. Sperm DNA fragmentation index, as measured by sperm chromatin dispersion, might not predict assisted reproductive outcome. Taiwan. J. Obstet. Gynecol. 57, 493–498 (2018).
Simon, L. et al. damage output parameters measured by the alkaline Comet assay and their importance. Andrologia 49, 1–12 (2017).
Nasr-Esfahani, M. H. et al. Effect of sperm DNA damage and sperm protamine deficiency on fertilization and embryo development post-ICSI. Reprod. Biomed. Online 11, 198–205 (2005).
Ribas-Maynou, J. et al. Alkaline and neutral Comet assay profiles of sperm DNA damage in clinical groups. Hum. Reprod. 27, 652–658 (2012).
Ribas-Maynou, J. et al. Double stranded sperm DNA breaks, measured by comet assay, are associated with unexplained recurrent miscarriage in couples without a female factor. PLoS ONE 7, e44679 (2012).
Langie, S. A. S., Azqueta, A. & Collins, A. R. The comet assay: past, present, and future. Front. Genet. 6, 266 (2015).
Garolla, A. et al. High-power microscopy for selecting spermatozoa for ICSI by physiological status. Reprod. Biomed. Online 17, 610–616 (2008).
Utsuno, H., Oka, K., Yamamoto, A. & Shiozawa, T. Evaluation of sperm head shape at high magnification revealed correlation of sperm DNA fragmentation with aberrant head ellipticity and angularity. Fertil. Steril. 99, 1573–1580.e1 (2013).
Nixon, B. et al. The role of the molecular chaperone heat shock protein A2 (HSPA2) in regulating human sperm-egg recognition. Asian J. Androl. 17, 568 (2015).
Inoue, N., Hagihara, Y., Wright, D., Suzuki, T. & Wada, I. Oocyte-triggered dimerization of sperm IZUMO1 promotes sperm–egg fusion in mice. Nat. Commun. 6, 8858 (2015).
Hiramoto, Y. & Baba, S. A. A quantitative analysis of flagellar movement in echinoderm spermatozoa. J. Exp. Biol. 76, 85–104 (1978).
Saggiorato, G. et al. Human sperm steer with second harmonics of the flagellar beat. Nat. Commun. 8, 1415 (2017).
Hansen, J., Rassmann, S., Jikeli, J. & Wachten, D. SpermQ–A simple analysis software to comprehensively study flagellar beating and sperm steering. Cells 8, 10 (2018).
Gallagher, M. T., Cupples, G., Ooi, E. H., Kirkman-Brown, J. C. & Smith, D. J. Rapid sperm capture: high-throughput flagellar waveform analysis. Hum. Reprod. 34, 1173–1185 (2019).
Di Caprio, G. et al. Quantitative label-free animal sperm imaging by means of digital holographic microscopy. IEEE J. Sel. Top. Quantum Electron. 16, 833–840 (2010).
Di Caprio, G. et al. 4D tracking of clinical seminal samples for quantitative characterization of motility parameters. Biomed. Opt. Express 5, 690 (2014).
Su, T.-W., Xue, L. & Ozcan, A. High-throughput lensfree 3D tracking of human sperms reveals rare statistics of helical trajectories. Proc. Natl Acad. Sci. USA 109, 16018–16022 (2012).
Daloglu, M. U. et al. Label-free 3D computational imaging of spermatozoon locomotion, head spin and flagellum beating over a large volume. Light Sci. Appl. 7, 17121 (2018).
Daloglu, M. U. et al. 3D imaging of sex-sorted bovine spermatozoon locomotion, head spin and flagellum beating. Sci. Rep. 8, 15650 (2018).
Dubey, V. et al. Partially spatially coherent digital holographic microscopy and machine learning for quantitative analysis of human spermatozoa under oxidative stress condition. Sci. Rep. 9, 3564 (2019).
de Wagenaar, B. et al. Microfluidic single sperm entrapment and analysis. Lab. Chip 15, 1294–1301 (2015).
de Wagenaar, B. et al. Spermometer: electrical characterization of single boar sperm motility. Fertil. Steril. 106, 773–780.e6 (2016).
You, J. B. et al. Live sperm trap microarray for high throughput imaging and analysis. Lab. Chip 19, 815–824 (2019).
Goodson, S. G. et al. CASAnova: a multiclass support vector machine model for the classification of human sperm motility patterns†. Biol. Reprod. 97, 698–708 (2017).
Goodson, S. G., Zhang, Z., Tsuruta, J. K., Wang, W. & O’Brien, D. A. Classification of mouse sperm motility patterns using an automated multiclass support vector machines model1. Biol. Reprod. 84, 1207–1215 (2011).
Hicks, S. A. et al. Machine learning-based analysis of sperm videos and participant data for male fertility prediction. Sci. Rep. 9, 16770 (2019).
Somasundaram, D. & Nirmala, M. Faster region convolutional neural network and semen tracking algorithm for sperm analysis. Comput. Methods Prog. Biomed. 200, 105918 (2021).
Sutskever, I., Vinyals, O. & Le, Q. V. Sequence to sequence learning with neural networks. Proc. Int. Conf. Neural Inform. Process. Syst. 2, 3104–3112 (2014).
Cho, K. et al. Learning phrase representations using RNN encoder–decoder for statistical machine translation. Proc. Conf. Empir. Methods Nat. Lang. Process. 28, 1724–1734 (2014).
Ng, J. Y. H. et al. Beyond short snippets: deep networks for video classification (IEEE, 2015).
Yao, L. et al. Describing videos by exploiting temporal structure (IEEE, 2015).
Donahue, J. et al. Long-term recurrent convolutional networks for visual recognition and description. IEEE Trans. Pattern Anal. Mach. Intell. 39, 677–691 (2017).
Kimmel, J., Brack, A. & Marshall, W. Deep convolutional and recurrent neural networks for cell motility discrimination and prediction. IEEE/ACM Trans. Comput. Biol. Bioinforma. 18, 562–574 (2019).
Liu, Q. et al. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 10, 2449 (2019).
Zhou, X. et al. Hybrid generative-discriminative learning for online tracking of sperm cell. Neurocomputing 208, 218–224 (2016).
Karahan, Ş. et al. How image degradations affect deep CNN-based face recognition? (IEEE, 2016).
Dodge, S. & Karam, L. Understanding how image quality affects deep neural networks (IEEE, 2016).
Teixeira, D. M. et al. Regular (ICSI) versus ultra-high magnification (IMSI) sperm selection for assisted reproduction. Cochrane Database Syst. Rev. 7, CD010167 (2013).
Ching, T. et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 15, 20170387 (2018).
Dai, C. et al. Automated non-invasive measurement of single sperm’s motility and morphology. IEEE Trans. Med. Imaging 37, 2257–2265 (2018).
Wainberg, M., Merico, D., Delong, A. & Frey, B. J. Deep learning in biomedicine. Nat. Biotechnol. 36, 829–838 (2018).
Intasqui, P., Agarwal, A., Sharma, R., Samanta, L. & Bertolla, R. P. Towards the identification of reliable sperm biomarkers for male infertility: a sperm proteomic approach. Andrologia 50, e12919 (2018).
Gijsberts, C. M. et al. Race/ethnic differences in the associations of the Framingham risk factors with carotid IMT and cardiovascular events. PLoS ONE 10, e0132321 (2015).
Buolamwini, J. & Gebru, T. Gender shades: intersectional accuracy disparities in commercial gender classification. Proc. Mach. Learn. Res. 81, 77–91 (2018).
Balki, I. et al. Sample-size determination methodologies for machine learning in medical imaging research: a systematic review. Can. Assoc. Radiol. J. 70, 344–353 (2019).
Vayena, E., Blasimme, A. & Cohen, I. G. Machine learning in medicine: addressing ethical challenges. PLoS Med. 15, e1002689 (2018).
Froomkin, A. M., Kerr I. & Pineau, J. in Called to Holiness 167–170 (Catholic Univ. America Press, 2017).
Price, W. N., Gerke, S. & Cohen, I. G. Potential liability for physicians using artificial intelligence. JAMA 322, 1765 (2019).
Reddy, S., Allan, S., Coghlan, S. & Cooper, P. A governance model for the application of AI in health care. J. Am. Med. Inform. Assoc. 27, 491–497 (2020).
Kuner, C., Svantesson, D. J. B., Cate, F. H., Lynskey, O. & Millard, C. Machine learning with personal data: is data protection law smart enough to meet the challenge? Int. Data Priv. Law 7, 1–2 (2017).
Richens, J. G., Lee, C. M. & Johri, S. Improving the accuracy of medical diagnosis with causal machine learning. Nat. Commun. 11, 3923 (2020).
Myszczynska, M. A. et al. Applications of machine learning to diagnosis and treatment of neurodegenerative diseases. Nat. Rev. Neurol. 16, 440–456 (2020).
Goecks, J., Jalili, V., Heiser, L. M. & Gray, J. W. How machine learning will transform biomedicine. Cell 181, 92–101 (2020).
Willemink, M. J. et al. Preparing medical imaging data for machine learning. Radiology 295, 4–15 (2020).
Alexander, A., Jiang, A., Ferreira, C. & Zurkiya, D. An intelligent future for medical imaging: a market outlook on artificial intelligence for medical imaging. J. Am. Coll. Radiol. 17, 165–170 (2020).
VerMilyea, M. et al. Development of an artificial intelligence-based assessment model for prediction of embryo viability using static images captured by optical light microscopy during IVF. Hum. Reprod. 35, 770–784 (2020).
Nayot, D., Bharti, R. M., Meriano, J. & Krivoi, A. Methods and systems for determining quality of an oocyte. US Patent 10,552,957 B2 (2020).
Char, D. S., Shah, N. H. & Magnus, D. Implementing machine learning in health care — addressing ethical challenges. N. Engl. J. Med. 378, 981–983 (2018).
Wijesekera, T. P., Sanders, L. & Windish, D. M. Reflections on diagnosis and diagnostic errors: a survey of internal medicine resident and attending physicians. J. Gen. Intern. Med. 35, 614–615 (2020).
Currie, G., Hawk, K. E., Rohren, E., Vial, A. & Klein, R. Machine learning and deep learning in medical imaging: intelligent imaging. J. Med. Imaging Radiat. Sci. 50, 477–487 (2019).
Giger, M. L. Machine learning in medical imaging. J. Am. Coll. Radiol. 15, 512–520 (2018).
Senders, J. T. et al. An introduction and overview of machine learning in neurosurgical care. Acta Neurochir. 160, 29–38 (2018).
Staartjes, V. E. et al. Machine learning in neurosurgery: a global survey. Acta Neurochir. 162, 3081–3091 (2020).
Pennig, L. et al. Primary central nervous system lymphoma: clinical evaluation of automated segmentation on multiparametric MRI using deep learning. J. Magn. Reson. Imaging 53, 259–268 (2021).
Schelb, P. et al. Classification of cancer at prostate MRI: deep learning versus clinical PI-RADS assessment. Radiology 293, 607–617 (2019).
Azencott, C.-A. Machine learning and genomics: precision medicine versus patient privacy. Phil. Trans. R. Soc. A 376, 20170350 (2018).
Tran, D., Cooke, S., Illingworth, P. J. & Gardner, D. K. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum. Reprod. 34, 1011–1018 (2019).
Horvitz, E. & Mulligan, D. Data, privacy, and the greater good. Science 349, 253–255 (2015).
Dardikman-Yoffe, G., Mirsky, S. K., Barnea, I. & Shaked, N. T. High-resolution 4-D acquisition of freely swimming human sperm cells without staining. Sci. Adv. 6, eaay7619 (2020).
Keys, R. Cubic convolution interpolation for digital image processing. IEEE Trans. Acoust. 29, 1153–1160 (1981).
Duchon, C. E. Lanczos filtering in one and two dimensions. J. Appl. Meteorol. 18, 1016–1022 (1979).
Qinlan, X., Hong, C. & Huimin, C. Improved example-based single-image super-resolution (IEEE, 2010).
Sert, E., Özyurt, F. & Doğantekin, A. A new approach for brain tumor diagnosis system: single image super resolution based maximum fuzzy entropy segmentation and convolutional neural network. Med. Hypotheses 133, 109413 (2019).
Ravì, D., Szczotka, A. B., Shakir, D. I., Pereira, S. P. & Vercauteren, T. Effective deep learning training for single-image super-resolution in endomicroscopy exploiting video-registration-based reconstruction. Int. J. Comput. Assist. Radiol. Surg. 13, 917–924 (2018).
Shaker, F. Human sperm head morphology data set (HuSHeM). Mendeley Data https://doi.org/10.17632/tt3yj2pf38.1 (2017).
Agarwal, A., Gupta, S. & Sharma, R. (eds) in Andrological Evaluation of Male Infertility 181–203 (Springer, 2016).
Adiga, S. K. & Kalthur, G. in Male Infertility (Gunasekaran, K. & Pandiyan, N.) 155–165 (Springer, 2017).
Tandara, M. et al. Sperm DNA integrity testing: big halo is a good predictor of embryo quality and pregnancy after conventional IVF. Andrology 2, 678–686 (2014).
Cortés-Gutiérrez, E. I., Dávila-Rodríguez, M. I. & López-Fernández, C. in A Clinician’s Guide to Sperm DNA and Chromatin Damage (eds Zini, A. & Agarwal, A.) 119–135 (Springer, 2018).
Bukatin, A., Kukhtevich, I., Stoop, N., Dunkel, J. & Kantsler, V. Bimodal rheotactic behavior reflects flagellar beat asymmetry in human sperm cells. Proc. Natl Acad. Sci. USA 112, 15904–15909 (2015).
Frimat, J.-P. et al. Make it spin: individual trapping of sperm for analysis and recovery using micro-contact printing. Lab. Chip 14, 2635 (2014).
Mathews, S. C. et al. Digital health: a path to validation. NPJ Digit. Med. 2, 38 (2019).
Jordan, M. I. & Mitchell, T. M. Machine learning: trends, perspectives, and prospects. Science 349, 255–260 (2015).
Yu, K.-H., Beam, A. L. & Kohane, I. S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2, 719–731 (2018).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Riordon, J., Sovilj, D., Sanner, S., Sinton, D. & Young, E. W. K. Deep learning with microfluidics for biotechnology. Trends Biotechnol. 37, 310–324 (2019).
Webb, S. Deep learning for biology. Nature 554, 555–557 (2018).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
Badrinarayanan, V., Kendall, A. & Cipolla, R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 2481–2495 (2017).
Yu, Y., Si, X., Hu, C. & Zhang, J. A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput. 31, 1235–1270 (2019).
Acknowledgements
This work was supported by the Collaborative Health Research Project funded by the Canadian Institute of Health Research (CIHR) and the Natural Sciences and Engineering Research Council of Canada (NSERC). R.N. acknowledges support from the Australian Research Council Discovery Program (DP190100343). The authors also gratefully acknowledge the Canada Research Chairs Program (DS). The authors thank K. Jarvi, T. Hannam and A. Lagunov for insightful discussions on the potential for machine learning in clinical applications.
Review criteria
Articles for this Review were selected based on the relevance to the topics discussed in the manuscript, namely sperm morphology assessment, DNA integrity, sperm motility and machine learning. Articles of interest were those describing, evaluating and discussing sperm morphology and DNA integrity assessment techniques, as well as those that propose the use of machine learning for sperm classification. As most current sperm morphology and DNA assessment approaches are based on techniques developed several decades ago, reports with publication from 1978 to 2020 have been referenced in the Review. In the ethics section, we have referenced specialized perspectives and review papers discussing the ethical issues of artificial intelligence and machine learning written by experts in legal, medical and technological sectors.
Author information
Authors and Affiliations
Contributions
J.B.Y., C.M., and Y.W. researched data for the manuscript, J.B.Y., J.R. and D.S. made substantial contributions to discussions of content. All authors wrote, reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
You, J.B., McCallum, C., Wang, Y. et al. Machine learning for sperm selection. Nat Rev Urol 18, 387–403 (2021). https://doi.org/10.1038/s41585-021-00465-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41585-021-00465-1
This article is cited by
-
The prospect of artificial intelligence to personalize assisted reproductive technology
npj Digital Medicine (2024)
-
Advances in microfluidic technology for sperm screening and in vitro fertilization
Analytical and Bioanalytical Chemistry (2024)
-
Frequency, morbidity and equity — the case for increased research on male fertility
Nature Reviews Urology (2024)
-
Basic, translational and clinical studies in reproductive medicine and clinical reproductive sciences
Journal of Translational Medicine (2023)
-
Artificial intelligence in academic writing: a paradigm-shifting technological advance
Nature Reviews Urology (2023)