Abstract
The retinoic acid-inducible gene I (RIG-I) receptor senses cytoplasmic viral RNA and activates type I interferons (IFN-I) and downstream antiviral immune responses. How RIG-I binds to viral RNA and how its activation is regulated remains unclear. Here, using IFI16 knockout cells and p204-deficient mice, we demonstrate that the DNA sensor IFI16 enhances IFN-I production to inhibit influenza A virus (IAV) replication. IFI16 positively upregulates RIG-I transcription through direct binding to and recruitment of RNA polymerase II to the RIG-I promoter. IFI16 also binds to influenza viral RNA via its HINa domain and to RIG-I protein with its PYRIN domain, thus promoting IAV-induced K63-linked polyubiquitination and RIG-I activation. Our work demonstrates that IFI16 is a positive regulator of RIG-I signalling during influenza virus infection, highlighting its role in the RIG-I-like-receptor-mediated innate immune response to IAV and other RNA viruses, and suggesting its possible exploitation to modulate the antiviral response.
Similar content being viewed by others
Main
Influenza virus is one of the most important causes of respiratory tract infection, resulting in at least 26 million influenza illnesses, 250,000 hospitalizations and 14,000 deaths during the 2019–2020 season in the United States (https://www.cdc.gov/flu/season/index.html). A(H1N1)/pdm2009, H3N2 and influenza B (B/Yamagata or B/Victoria lineage) viruses are mainly responsible for seasonal influenza epidemics each year. Moreover, avian influenza virus remains a notable additional threat to human health, in particular the H5, H7 and H9 subtypes. Although vaccination is the most effective way to control influenza A viruses (IAVs), prediction of evolving immunogenic epitopes as well as challenges in vaccine production and distribution often limit vaccine efficacy and availability. Furthermore, mutations in antiviral-resistant IAV strains continue to be identified, including the H274Y mutation in neuraminidase (NA)1,2 and three major mutations (L26F, V27A and S31N) in the M2 protein3,4. Hence, the development of effective interventions against influenza virus infection remains an outstanding public health need. Targeting an essential host factor critical for influenza infection is a promising antiviral strategy5.
Several families of pattern recognition receptors have been described, including Toll-like receptors, retinoic acid inducible gene-I (RIG-I)-like receptors (RLRs) and nucleotide-oligomerization-domain-like receptors, which are involved in the recognition of influenza viruses6,7,8. Among these, RIG-I is thought to be the most important sensor of influenza virus9; it binds the 5′ppp-RNA of the virus, leading it to undergo conformational changes and exposing its caspase activation and recruitment domains10, which is then ubiquitinated by the action of E3 ligases such as tripartite motif 25 (TRIM25)11 and RIPLET12. TRIM25 activates RIG-I through the generation of unanchored K63-linked polyubiquitin chains interacting with the caspase activation and recruitment domains13 or through the generation of anchored K63-linked polyubiquitin chains attached to K172 of RIG-I11. This process results in the interaction of RIG-I with the mitochondrial antiviral signalling (MAVS) adaptor, which leads to the subsequent activation of IRF3, IRF7 and NF-κB and thereby inducing the expression of type I interferon (IFN-I) and pro-inflammatory cytokines14. Therefore, RIG-I activation has to be tightly regulated to ensure effective virus inhibition while minimizing an excessive inflammatory response.
IFN-γ-inducible protein-16 (IFI16) is a member of the pyrin and HIN domain (PYHIN)-containing protein family, which encodes a class of homologous proteins that share a 200-amino acid signature motif (HIN)15. IFI16 was first reported as a sensor of transfected and viral DNA involved in innate signalling16,17, and it functions as an innate immune sensor in eukaryotic cells18,19,20,21,22,23,24. IFI16 senses double-stranded DNA from invading DNA viruses, including herpes simplex virus 1, Kaposi sarcoma-associated herpesvirus and vaccinia virus21,24,25, single-stranded DNA from HIV-infected CD4+ T cells and nuclear-damaged DNA from etoposide-treated keratinocytes22,26. DNA recognition by IFI16 induces activation of the stimulator of interferon genes–TANK-binding kinase 1–interferon regulatory factor 3 (STING–TBK1–IRF3) pathway, which leads to the induction of IFN-I or the ASC–caspase 1-dependent inflammasome to produce interleukin-1β (IL-1β)21,24,26. Additionally, a role for IFI16 in RNA virus infection has been identified, whereby IFI16 transcriptionally regulates the gene expression of IFN-I in Sendai virus infection27. The murine orthologue of IFI16 (p204) is highly induced in mouse hepatitis coronavirus infection and inhibits IRF7-mediated IFN-I production28. Moreover, it was shown that IFI16 interacts with MAVS to promote the MAVS-mediated production of IFN-I that inhibits porcine reproductive and respiratory syndrome virus 2 replication29. More recently, IFI16 was found to directly bind chikungunya virus genome RNA and restrict viral replication and maturation30, which further suggests that IFI16 may play a crucial role in RNA virus infection. However, the precise role of IFI16 in influenza virus infection has not been elucidated.
Here, we show that IFI16, which was identified as an influenza viral RNA-binding protein (RBP), is highly induced both in vitro and in vivo during IAV infection. We further demonstrate that IFI16 upregulates RIG-I transcription by binding its HINa domain to the RIG-I promoter, and interacts with both IAV viral RNA (vRNA) and RIG-I to promote influenza-virus-induced K63-linked polyubiquitination of RIG-I. Collectively, these results indicate that IFI16 is a key positive regulator of RIG-I signalling in antiviral innate immune responses to influenza virus infection.
Results
IFI16 is a viral RBP involved in influenza virus infection
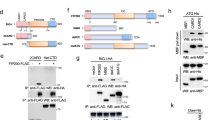
Owing to its RNA genome, influenza virus utilizes RBPs of both viral and host origin for its replication. To comprehensively uncover vRNA–host protein interactions, we performed affinity purification coupled with mass spectrometry (AP–MS) analysis of influenza vRNA complexes. The eight vRNA segments of H7N9 virus were individually transcribed and labelled with biotin in vitro and then incubated with lysates from IAV-infected THP-1 cells. vRNA complexes bound to streptavidin magnetic beads were analysed by MS (Fig. 1a). Domain-enrichment analysis of co-purified proteins for each RNA bait revealed that more than 90% of the isolated proteins harboured nucleic-acid-binding domains (Fig. 1b). We identified 70 candidate viral RBPs that could bind to no fewer than 3 baits. A high number of co-purified proteins have been reported as RBPs, such as PTBP1 (ref. 31), TRA2B32, DDX5 (ref. 33), DDX3X34 and RBBP6 (ref. 35), which indicates that our approach was effective in identifying vRNA-interacting proteins for influenza virus. The expression results demonstrated that IFI16 was most highly expressed for the duration of IAV infection (Fig. 1c), which suggests that IFI16 may play an important role in IAV infection.
a, Schematic representation of the vRNA pull-down assay and MS approach to identify viral RBPs in THP-1 cells. b, Protein domain types significantly enriched among the human interacting proteins with indicated vRNA baits. The value range of Q value is [0,1]. The closer Q is to zero, the more significant the enrichment. Domains with Q ≤ 0.05 are defined as domains that are significantly enriched in differentially expressed genes. c, mRNA expression levels, determined by RT–qPCR, of 70 candidate viral RBPs in THP-1 cells infected with PR8 virus at 2.0 m.o.i. RNA samplings were taken at 0, 6, 12 and 24 h.p.i. d, IFI16 protein expression in PR8-virus-infected (m.o.i. = 1). A549 cells were evaluated by western blotting at 0, 6, 12, 18 and 24 h.p.i. (β-actin detection was used as the loading control). e, Intracellular localization of IFI16 was assessed in PR8-virus-infected A549 cells at 0, 6, 12 and 18 h.p.i. by confocal microscopy. f, Quantification of the intracellular localization of IFI16 in cells as in e. The data represent the mean ± s.d. (n = 3 independent experiments). g, A549 cells were infected with PR8 virus at 0, 12 and 24 h.p.i. Whole cell lysate (WCL) was then immunoprecipitated with anti-IFI16. Bound proteins were analysed by immunoblotting with anti-acetylated lysine (Acet. Lys). h, A549 cells were pre-incubated with C-646 for 2 h, infected with PR8 virus for 1 h, washed with PBS and incubated in complete medium with or without C-646. In situ PLA assays were used to assess IFI16 acetylation with anti-IFI16 and anti-acetylated lysine antibodies at 24 h.p.i. For b, the results were assessed using parametric paired t-test (Student’s one-tailed t-test). For d–h, the data presented are representative of three independent experiments. Statistical significance in f was determined by unpaired two-tailed Student’s t-test.
To determine whether IFI16 is involved in IAV replication, we first evaluated IFI16 expression in PR8-virus-infected human cells. IFI16 mRNA levels were significantly upregulated in THP-1, A549 and HEK293 cells during PR8 infection (Extended Data Fig. 1a–c). We also found that PR8 infection significantly elevated p204 expression in mice lung tissues at 1 day post infection (d.p.i.) and at 3 and 5 d.p.i. (Extended Data Fig. 1d). PR8 virus actively induced the expression of IFI16 in THP-1 (Extended Data Fig. 1e) and A549 cells (Fig. 1d). Furthermore, ultraviolet-inactivated PR8 virus did not induce IFI16 expression in A549 cells compared with live virus infection (Extended Data Fig. 1f–g). Collectively, these findings suggest that IFI16 is involved in the modulation of influenza virus infection.
In uninfected cells, IFI16 was mainly localized in the nucleus, whereas PR8 infection induced an accumulation of IFI16 in the nucleus and cytoplasm (Fig. 1e,f). Consistent with the immunostaining results, PR8 virus infection caused a build-up of IFI16 protein in the cytoplasm and nucleus (Extended Data Fig. 1g). Moreover, treatment of A549 cells with human IFN-γ or polyinosinic:polycytidylic acid (poly(I:C)) induced IFI16 accumulation in the nucleus and cytoplasm (Extended Data Fig. 1i–k). It was reported that acetylation of IFI16 modulates its cellular distribution and cytoplasm translocation in DNA virus infection25,36. During IAV infection, we also detected the acetylation of IFI16 in PR8-virus-infected A549 cells at 12 and 24 hours post infection (h.p.i.) (Fig. 1g and Extended Data Fig. 1l). Additionally, we performed in situ proximity ligation assays (PLAs), and low levels of nuclear acetylated IFI16 dots were detected in uninfected A549 cells (Fig. 1h). In contrast, acetylated IFI16 dots were clearly elevated in the nucleus and cytoplasm at 24 h.p.i. (Fig. 1h). We found that the IAV-induced accumulation of IFI16 in the cytoplasm was abolished in C-646-treated cells (Extended Data Fig. 1m). Collectively, IFI16 is highly upregulated during IAV infection and its associated acetylation can confer functional modifications such as stabilization of the IFI16 protein.
IFI16 inhibits IAV infection in vitro and in vivo
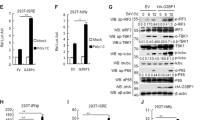
To determine the impact of IFI16 on influenza virus, IFI16–Flag vectors were transfected into A549 cells followed by PR8 infection at a multiplicity of infection (m.o.i.) of 1. Overexpression of IFI16 significantly reduced the viral titre of PR8 at 18 h.p.i. (Fig. 2a) and reduced the expression of viral NP and M1 proteins (Fig. 2b). Increasing the amounts of transfected IFI16–Flag vectors in HEK293T-Gluc cells resulted in impaired viral replication in a dose-dependent manner (Fig. 2c). In addition, overexpression of IFI16 inhibited the expression of mRNA and vRNA of NP and M1 genes in A549 cells (Extended Data Fig. 2a) and of protein levels of NP at 6, 12 and 18 h.p.i. in HEK293 cells (Extended Data Fig. 2b). Conversely, when IFI16 was knocked down by short interfering RNA (siRNA) in A549 cells, there were significant increases in viral protein and titres of progeny viruses (Extended Data Fig. 2c,d). To investigate the function of endogenous IFI16 during IAV infection, IFI16+/+ and IFI16−/− A549 cells were infected with PR8 virus. Infected IFI16−/− A549 cells produced higher viral titres at 12 and 24 h.p.i. (Fig. 2d) and protein levels of viral NP and M1 at 12 and 18 h.p.i. (Fig. 2e). In a gain-of-function experiment, exogenous expression of IFI16 in IFI16−/− A549 cells effectively reduced the expression of viral NP and M1 proteins at 12 and 18 h.p.i. (Fig. 2f). Accordingly, we found that the replication of green fluorescent protein (GFP)-tagged PR8 virus was markedly increased in IFI16−/− A549 cells compared with IFI16+/+ A549 cells (Fig. 2g). Collectively, these findings indicate that IFI16 inhibits influenza virus replication in human cells.
a, A549 cells were transfected with IFI16–Flag plasmids or control for 24 h and then infected with PR8 virus at a m.o.i. of 1 for 18 h. Viral titres were measured by TCID50 assays. Data are presented as the mean ± s.e.m. and are representative of three independent experiments. b, Left: A549 cells were transfected with IFI16–Flag plasmids or control for 24 h and then infected with PR8 virus at a m.o.i. of 1, and NP and M1 proteins were detected. Right: data were quantified and shown as the ratio of NP to β-actin and M1 to β-actin. The data represent the mean ± s.e.m. (n = 3). c, HEK293T-Gluc cells were transfected with different amounts of IFI16–Flag or control, followed by infection with PR8 virus (m.o.i. = 1) for 24 h. Viral infectivity was determined. Data are presented as the mean ± s.e.m. and are representative of three independent experiments. d, Viral titres in PR8-virus-infected (m.o.i. = 1) IFI16+/+ (IFI16 WT) and IFI16−/− (IFI16 KO) A549 cells were measured by TCID50 assay. Data are presented as the mean ± s.e.m. and are representative of three independent experiments. e, Left: the NP and M1 protein expression levels in PR8-virus-infected (1 m.o.i.) IFI16+/+ and IFI16−/− A549 cells were analysed. Right: data were quantified and shown as the ratio of NP to β-actin and M1 to β-actin. The data represent the mean ± s.e.m. (n = 3). f, Left: IFI16+/+ and IFI16−/− A549 cells were transfected with IFI16–Flag plasmids or empty control for 24 h and then infected with PR8 virus at a m.o.i. of 1. NP and M1 proteins in virus-infected A549 cells were detected. Right: data were quantified and shown as the ratio of NP to β-actin and M1 to β-actin. The data represent the mean ± s.e.m. (n = 3). g, Fluorescence microscopy images of viral replication (green) in IFI16+/+ and IFI16−/− A549 cells after infection with GFP–PR8 virus for 12 h. For b and d–g, data are representative of three independent experiments. Statistical significance in a–f was determined by unpaired two-tailed Student’s t-test.
Additionally, PR8-infected wild-type (WT) mice suffered significantly less weight loss than p204-deficient (p204−/−) mice, and they started to regain body weight by 8 d.p.i. (Fig. 3a). Moreover, survival rates of PR8 infected p204−/− mice were significantly poorer than infected WT mice (Fig. 3b), which suggests that control of IAV infection in vivo also requires p204. In lung tissues of p204−/− mice, PR8 virus replication at 4 and 6 d.p.i. was higher (Fig. 3c), with accompanying greater viral NP mRNA and vRNA levels (Fig. 3d), in lung tissues of p204−/− mice than in corresponding WT mice. Moreover, viral NP staining was more intense in lung sections of p204−/− mice at 5 d.p.i. than in corresponding WT mice (Fig. 3e). Notably, gross pathology and histopathology analyses revealed that lung tissues of PR8-virus-infected p204−/− mice displayed more extensive damage at 3 and 5 d.p.i. (Fig. 3f), and more severe inflammatory damage at 3 d.p.i. (Fig. 3g), than in corresponding WT mice. Accordingly, IL-6 and MCP-1 protein levels in bronchoalveolar fluid (BALF) were also significantly higher in PR8-infected WT mice than those found in corresponding p204−/− mice (Fig. 3h,i). Furthermore, p204 deficiency dramatically inhibited the transcription of Ifnb, Viperin, Oas1, Isg15 and Il6 in PR8-virus-infected bone marrow-derived macrophages (BMDMs) (Fig. 3j–n). Therefore, IFI16 is required for host defence against influenza virus infection in vitro and in vivo.
a, WT and p204−/− mice (n = 6) were infected with 50 TCID50 of PR8 virus. Changes in body weight were monitored daily. Data are presented as the mean ± s.d. b, Survival of WT and p204−/− mice infected with 50 TCID50 of PR8 virus. Data presented were pooled from three independent experiments. Kaplan–Meier survival curves were compared using log-rank (Mantel–Cox) analysis. c, Viral titres in lung tissues from 50 TCID50 of PR8-virus-infected WT and p204−/− mice at 4 and 6 d.p.i. were determined by EID50 assay. Data are from three independent experiments with n = 6 mice per group run in triplicate. Error bars indicate s.e.m. d, Viral NP mRNA (left) and vRNA (right) in lung tissues of PR8-virus-infected WT and p204−/− mice at 3 d.p.i. were determined by RT–qPCR. Data are presented as the mean ± s.e.m. and are representative of three independent experiments. e, Viral NP protein expression in lung tissue sections from virus-infected WT and p204−/− mice (n = 3) mice was examined by immunohistochemistry. Representative sections of one mouse out of three are shown. Scale bars, 800 μm (top) and 80 μm (bottom). f, Images of gross lesions (outlined in blue) of lung tissues from virus-infected WT and p204−/− mice at 3 and 5 d.p.i. Representative sections of one mouse out of three are shown. g, H&E staining of lung tissues from virus-infected WT and p204−/− mice at 3 d.p.i. Scale bars, 600 μm (left) and 120 μm (right). Representative H&E staining images are from six mice per group of three independent experiments. h,i, IL-6 and MCP-1 levels in BALF from virus-infected WT and p204−/− mice (n = 4) were quantified. Data are presented as the mean ± s.d. j–n, mRNA expression of Ifnb (j), viperin (k), Oas1 (l), Isg15 (m) and Il6 (n) in BMDMs of virus-infected WT and p204−/− mice was determined by RT–PCR. Data presented as mean ± s.e.m. and are representative of three independent experiments. Statistical significance in a, c, d and h–n was determined by unpaired two-tailed Student’s t-test.
IFI16 enhances the production of IFN-I in IAV infection
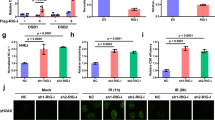
DNA recognition by IFI16 induces the activation of the STING–TBK1–IRF3 pathway, which leads to the induction of IFN-I21,24,26. To investigate the mechanism of innate immune activation by IFI16, we performed transcriptomics analysis in IFI16+/+ and IFI16−/− A549 cells infected with PR8 virus for 12 h. In the Gene Ontology (GO) term enrichment analysis, ten enriched terms identified were related to antiviral responses (Fig. 4a). The number of genes associated with individual terms and enrichment probability were lower in IFI16−/− A549 cells (Fig. 4a). Sixteen IFN-stimulated genes (ISGs) were exclusively upregulated in IFI16+/+ cells during IAV infection, while only 4 ISGs were upregulated in IFI16−/− A549 cells (Fig. 4b). Collectively, these results show that the induction of ISGs is reduced in the absence of IFI16. We next evaluated the function of IFI16 in the induction of IFN-I. Quantitative PCR with reverse transcription (RT–qPCR) analysis showed that overexpression of IFI16 enhanced PR8-virus-induced expression of IFNB, ISG15 and IL6 at 6 and 12 h.p.i. (Extended Data Fig. 3a–c). Conversely, IFI16 deficiency greatly reduced the PR8-virus-induced expression of IFNB, RIG-I, ISG56 and viperin (Fig. 4c–f) as well as IFNA4, IRF7 and CXCL5 (Extended Data Fig. 3d–f) in A549 cells. Furthermore, overexpression of IFI16 in A549 cells increased PR8-virus-induced IFN-β protein production at 12 and 18 h.p.i. in a dose-dependent manner (Fig. 4g), while IFI16 deficiency markedly decreased IFN-β production (Fig. 4h). These results suggest that IFI16 mediates antiviral effects through influenza-virus-induced IFN-I signalling.
a, GO term analysis of upregulated host proteins in IAV-infected IFI16+/+ and IFI16−/− A549 cells. The top ten GO terms are ordered by enrichment P values. The P values indicated in the legend and indices in cells correspond to the number of protein groups associated with individual GO terms in respective comparisons. b, Differential expression of host genes between IAV-infected and mock conditions in IFI16+/+ (x axis) and IFI16−/− (y axis). Proteins exhibiting significant differential responses are highlighted in green (ISGs) or orange (non-ISGs). c–f, Gene expression levels of IFNB (c), RIG-I (d), ISG56 (e) and Viperin (f) in PR8-virus-infected IFI16+/+ and IFI16−/− A549 cells at 0, 6 and 12 h.p.i. were determined by qPCR. g, A549 cells were transfected with IFI16–Flag expression vectors or empty vector (EV) for 24 h and then infected with PR8 virus. IFN-I levels in supernatants were quantified using IFN bioassays at 2, 12, 18, 24 and 36 h.p.i. at 1 m.o.i. h, IFI16+/+ and IFI16−/− A549 cells were infected with PR8 virus at 1 m.o.i. IFN-I levels in supernatants were then measured by IFN bioassays at 2, 12, 18, 24 and 36 h.p.i. i, A549 cells were transfected with IFI16–Flag expression vectors or empty control for 24 h and then infected with 1 m.o.i. of PR8 virus for 0, 2, 4, 8 and 12 h. RIG-I-triggered downstream signalling molecules were evaluated using the indicated antibodies. j, RIG-I-triggered downstream signalling molecules in PR8-virus-infected IFI16+/+ and IFI16−/− A549 cells at 0, 4, 8 and 12 h.p.i. were analysed using the indicated antibodies. k, IFI16+/+ and IFI16−/− A549 cells were infected with PR8 virus at 5 m.o.i. Nuclear localization of p-IRF3 was then determined by p-IRF3 (Ser396) intracellular immunostaining for confocal microscopy. l, Quantification of nuclear localization of p-IRF3 (Ser396). For i–k, data are representative of three independent experiments. For a, the results were assessed using parametric paired t-test (Student’s one-tailed t-test). For c–h and l, data are presented as the mean ± s.d. from three independent experiments, and the significance of the results was assessed using parametric paired t-test (Student’s two-tailed t-test).
To examine the contribution of IFI16 to RIG-I signalling, we transfected IFI16–Flag or control plasmids into A549 cells followed by infection with PR8 virus at 1 m.o.i. Overexpression of IFI16 in infected cells enhanced protein detection of RIG-I, phosphorylated TBK1 (p-TBK1), p-IRF3 and p-P65 relative to infected controls (Fig. 4i). In contrast, IFI16 deficiency in infected cells led to reduced protein detection of RIG-I, p-TBK1 and p-IRF3 compared with the IFI16+/+ group (Fig. 4j). Consistent with these results, overexpression of IFI16 in the presence of 5′ppp-RNA also increased protein detection of RIG-I, p-TBK1 and p-IRF3 (Extended Data Fig. 3g). Furthermore, exogenous expression of IFI16 in infected IFI16−/− A549 cells restored protein expression of RIG-I, p-TBK1 and p-IRF3 (Extended Data Fig. 3h). The immunofluorescence results showed that IFI16 deficiency reduced the level of p-IRF3 (Ser396) in the nucleus in PR8-virus-infected A549 cells (Fig. 4k,l). Taken together, these findings indicate that IFI16 is a potent stimulator of the IFN-I response in influenza virus infection.
IFI16 transcriptionally upregulates the expression of RIG-I
Overexpression of IFI16 progressively upregulated RIG-I protein expression in A549 cells in a time-dependent manner (Fig. 5a) and in a dose-dependent manner (Extended Data Fig. 4a). HEK293 cells co-transfected with IFI16–Myc and RIG-I–Flag followed by cycloheximide treatment did not exhibit evidence of RIG-I protein degradation (Extended Data Fig. 4b). Overexpression of IFI16 also increased the protein and mRNA expression of RIG-I in IFNAR1−/− A549 cells in a dose-dependent manner (Extended Data Fig. 4c,d). Furthermore, A549 cells transfected with IFI16–Flag vectors showed rising levels of RIG-I mRNA in a vector dose-dependent manner (Extended Data Fig. 4e) and in a progressive time-dependent manner (Fig. 5b). Consistent with the findings in A549 cells, mRNA expression of RIG-I in PR8-virus-infected lung tissues of p204-deficient mice was significantly reduced at 1 and 3 d.p.i. compared with corresponding WT mice (Extended Data Fig. 4f). Taken together, our results indicate that IFI16 upregulates RIG-I at the transcriptional level.
a, RIG-I expression in IFI16–Flag overexpressing A549 cells at 0, 18, 24 and 30 h post transfection was detected by western blotting. β-actin detection was used as the loading control. b, RIG-I mRNA expression in A549 cells transfected with IFI16–Flag plasmids for 0, 12, 18 and 24 h was quantified by RT–qPCR. c Luciferase activity levels of the RIG-I reporter in HEK293 cells transfected with increasing amounts of IFI16–Flag expression vectors for 24 h. Luciferase levels were normalized to Renilla levels. Values shown are fold changes over empty vector control. d, Schematic of the 2,000-bp promoter sequence of RIG-I and corresponding mutants. e, HEK293 cells were co-transfected with the RIG-I promoter mutant constructs and increasing amounts of IFI16–Flag as indicated. Luciferase levels were normalized to Renilla values. Values are shown as the fold-change over the empty vector control. f,g, Schematic of part of the RIG-I promoter sequence and every 50-bp deletion mutants (f), and a luciferase reporter assay (g) on the truncated mutants was performed as described in e. h,i, Schematic of part of the RIG-I promoter sequence and every 10-bp deletion mutants (h), and a luciferase reporter assay (i) of these truncated mutants was performed as described in e. j, A549 cells were infected with PR8 virus for 12 h followed by ChIP assay. The promoter sequence binding to IFI16 was determined by RT–qPCR. k, IFI16+/+ and IFI16−/− A549 cells were infected with PR8 virus for 0 and 12 h, followed by ChIP assay. RNA Pol II recruitment to the RIG-I promoter was assessed by RT–qPCR. Data are presented as percent input minus IgG background. l, Luciferase activity of the RIG-I promoter-luciferase reporter in HEK293 cells transfected with full-length IFI16 vectors and truncated mutant plasmids. Luciferase levels were normalized to Renilla values. Values are shown as the fold-change over the empty vector control. For a, data are representative of three independent experiments. For b–l, data are presented as the mean ± s.d. from three independent experiments. Statistical significance in b, c and e–l was determined by unpaired two-tailed Student’s t-test.
To further assess the role of IFI16 in the regulation RIG-I, a luciferase reporter vector driven by the RIG-I promoter was used in transfections of HEK293 cells. The RIG-I promoter was responsive to IFI16 overexpression in a vector dose-dependent manner (Fig. 5c), which indicates that IFI16 is capable of transactivating the RIG-I promoter.
To determine the minimum promoter length of RIG-I that is responsive to IFI16 induction, an extensive series of RIG-I promoter deletion constructs, each spliced to the luciferase reporter gene, was generated for co-transfections with increasing amounts of IFI16–Flag expression vectors (Fig. 5d–i). The minimum RIG-I promoter responsive to IFI16–Flag overexpression was between −371 and −360 bp in length (Fig. 5i). Furthermore, Flag antibody pull-down experiments, based on a series of biotinylated double-stranded DNA probes that spanned the minimum promoter section (Extended Data Fig. 4g), showed that only probe p2-mut4, which harboured mutations between the −371 and −360 region of the promoter, failed to bind IFI16–Flag (Extended Data Fig. 4h). Chromatin immunoprecipitation (ChIP) and qPCR assays, using two sets of primer pairs that target the −500 to −250 bp region of the RIG-I promoter, further demonstrated direct binding of IFI16 to the RIG-I promoter (Fig. 5j). Collectively, these results show that IFI16 is capable of binding to the RIG-I promoter to promote RIG-I transcription.
Next, to determine whether IFI16 affects the recruitment of RNA polymerase II (Pol II) to the RIG-I promoter, ChIP was performed with an anti-Pol II antibody on IFI16+/+ and IFI16−/− A549 cells infected with PR8 virus at 1 m.o.i. for 0 and 12 h. qPCR targeting the detection of the basal promoter site of RIG-I showed more RNA Pol II binding to the basal promoter of infected IFI16+/+ than infected IFI16−/− A549 cells (Fig. 5k), but no significant difference for the GAPDH promoter (Extended Data Fig. 4i), which suggests that during infection, IFI16 facilitates RNA Pol II recruitment to the basal promoter of RIG-I.
To investigate the domain(s) in IFI16 responsible for activating RIG-I transcription, we constructed IFI16 expression mutants bearing different domain deletions (Extended Data Fig. 4j). Mutants without the HINa domain completely lost the ability to activate the RIG-I–luc reporter gene (Fig. 5l). In a separate experiment, we also found that in the absence of the HINa domain, there was no binding of IFI16 to a biotinylated RIG-I promoter (Extended Data Fig. 4k). Deletion of the HINa domain in IFI16 abrogated its antiviral activity in response to PR8 virus infection (Extended Data Fig. 4l). Taken together, these data show that IFI16 binds the RIG-I promoter with its HINa domain facilitating the recruitment of RNA Pol II to the site, thereby enhancing the transcriptional activation of RIG-I.
IFI16 binds vRNAs and interacts with RIG-I
Since IFI16 is also a RBP, we determined whether IFI16 binds vRNAs as with the RIG-I promoter. Pull-down assays using cell lysates of HEK293 cells, separately overexpressing Flag-tagged IFI16, RIG-I, MDA-5 and TBK1 proteins, incubated with biotinylated NP vRNA showed binding of IFI16, RIG-I and MDA-5 to the vRNAs (Extended Data Fig. 5a). Microscale thermophoresis technology (MST) results showed that the HINa domain also exhibited high affinity for full-length NS vRNA (Extended Data Fig. 5b). To further demonstrate binding between IFI16 and other IAV RNAs, purified glutathione S-transferase (GST)–IFI16 proteins were incubated with fluorescein-labelled HA, NP, PA and PB2 vRNAs. MST assays also showed IFI16 binding with the different vRNAs (Extended Data Fig. 5c). Furthermore, RNA fluorescence in situ hybridization (FISH) analysis revealed increasing colocalization of IFI16 with NP vRNA in PR8-virus-infected A549 cells at 6 and 12 h.p.i. (Fig. 6a). To identify bound vRNAs during infection, RNA eluted from co-precipitation of IFI16–Flag from IAV-infected RIG-I−/− cells was subjected to deep sequencing analysis, which detected IFI16 binding to all eight IAV gene segments (Fig. 6b). RIG-I has been found with IAV genomic fragments37. IFI16 was also specifically enriched in genomic IAV segments during infection (Fig. 6b). Importantly, only WT IFI16 and IFI16 mutants bearing the HINa domain associated with vRNA fragments during infection (Fig. 6c and Extended Data Fig. 5d,e). Taken together, these results indicate that during infection, IFI16 can directly bind viral genome RNAs via its HINa domain.
a, Images showing colocalization of endogenous IFI16 (green) and viral NP RNA (red) in PR8-virus-infected A549 cells at 0, 6 and 12 h.p.i. was detected by RNA FISH. Nuclei were stained with DAPI (blue). Insets: quantification of the colocalization of IFI16 and NP RNA in cells (bar plots). Data represent the mean ± s.d. from three biological samples. b, Integrated Genome Viewer representation of captured IAV genomic (negative polarity) reads from Flag IPs of PR8-infected RIG-I−/− cells overexpressing IFI16–Flag (top) or RIG-I–Flag (bottom). Each horizontal green bar represents a single 150-nt read and the position where it aligns relative to an IAV gene segment. c, PCR detection of PB2 vRNAs in eluted RNA from RIG-I–Flag, IFI16–Flag and indicated Flag-tagged IFI16 deletions. d, A549 cells were infected with PR8 virus at 5 m.o.i. for the indicated duration, followed by co-IP with anti-IFI16 or IgG and immunoblotting analysis with anti-RIG-I antibody. e, A549 cells were infected with PR8 virus at 5 m.o.i. for the indicated duration, followed by co-IP with anti-RIG-I or IgG and immunoblotting analysis with anti-IFI16 antibody. f, A549 cells were infected with 1 m.o.i. of PR8 virus and then analysed by PLA with anti-IFI16 and anti-RIG-I antibodies at 0, 12 and 24 h.p.i. The right panels are enlarged. Red points indicate IFI16 plus RIG-I complexes, green points indicate the viral M1 protein. Data are representative of three independent experiments.
To determine whether IFI16 is involved in the recognition of vRNAs by RIG-I during influenza virus infection, co-immunoprecipitation (co-IP) assays were performed, and the results demonstrated that there is an endogenous interaction of IFI16 and RIG-I during PR8 virus infection in A549 cells (Fig. 6d,e). Furthermore, in situ PLA microscopy showed colocalization of IFI16 and RIG-I in the cytoplasm of PR8-virus-infected A549 cells (Fig. 6f). Consistent with the PLA results, confocal microscopy also found an increasing interaction between IFI16 and RIG-I during IAV infection (Extended Data Fig. 5f). The PYRIN domain can mediate the interaction between IFI16 and host proteins38,39. Accordingly, the IFI16-ΔPYRIN–GFP mutant was unable to colocalize with RIG-I in HEK293 cells, thereby indicating that the PYRIN domain is required in the protein–protein interaction (Extended Data Fig. 5g). Interestingly, PYRIN–GFP transfection into HEK293 cells resulted in its protein colocalization with RIG-I, which formed filamentous structures (Extended Data Fig. 5h,i). IFI16 filamentous structures are involved in antiviral responses by associating with host restriction factors40,41,42. Collectively, these findings indicate that IFI16 binds influenza vRNAs and associates with RIG-I in influenza-virus-infected cells.
RIG-I is required for IFI16-mediated antiviral responses in IAV infection
K63-linked polyubiquitination of RIG-I by TRIM25 is essential for its activation11. To explore the potential role of IFI16 in the promotion of RIG-I signalling, we examined the interaction between TRIM25 and RIG-I in PR8-virus-infected IFI16+/+ and IFI16−/− A549 cells. Co-IP and in situ PLA experiments indicated that IFI16 deficiency impaired the interaction and colocalization between RIG-I and TRIM25 due to their reduced expression in infected IFI16−/− cells, and inhibited K63-linked ployubiquitination of RIG-I during virus infection (Extended Data Figs. 6a,b and 7a). To further determine the impact of IFI16 on RIG-I polyubiquitination, we transfected HEK293 cells with Myc control or IFI16–Myc, RIG-I–Flag, TRIM25–His and either HA-ubiquitin, HA-ubiquitin-K48 or HA-ubiquitin-K63 plasmids. Co-IP experiments showed that IFI16 remarkably enhanced the interaction between TRIM25 and RIG-I and promoted RIG-I polyubiquitination in the presence of HA-ubiquitin and HA-ubiquitin-K63, but not HA-ubiquitin-K48 (Extended Data Fig. 7b). Thus, IFI16 facilitates TRIM25 binding to RIG-I and consequently promotes K63-linked polyubiquitination of RIG-I. RNA immunoprecipitation (RIP) and RT–qPCR assays showed that overexpression of IFI16 significantly enhanced the RIG-I binding to PA vRNA (Extended Data Fig. 7c). Correspondingly, IFI16 deficiency also markedly reduced RIG-I binding to PA vRNA in PR8-virus-infected A549 cells (Extended Data Fig. 7d). RIP and electrophoretic mobility shift assays (EMSAs) and RNA pull-down experiments further demonstrated that the binding of PA vRNA with RIG-I is sharply increased with increasing amounts of IFI16 (Extended Data Fig. 6c,d). Furthermore, RNA co-purified with RIG-I by RIP analysis from IAV-infected IFI16+/+ cells induced the production of IFN-β more effectively than that from IFI16−/− cells (Extended Data Fig. 7e,f), which suggests that IFI16 facilitates the stable binding of RIG-I to vRNAs during IAV infection. Importantly, only WT IFI16 and IFI16 mutants bearing the HINa and PYRIN domain, but not HINb, promoted vRNA binding to RIG-I (Extended Data Fig. 6e) and enhanced the production of IFN-β during IAV infection (Extended Data Fig. 6f). Consistent with these observations, IFN-β reporter assays showed that overexpression of IFI16 significantly enhanced PR8-virus-induced expression of IFN-β–luciferase reporter genes (Extended Data Fig. 7g). Moreover, the RIG-I-induced activity of IFN-β–luciferase reporter rose with increasing amounts of IFI16 expression vectors used in transfections (Extended Data Fig. 7h). Taken together, these results suggest that IFI16 promotes the activation of RIG-I signalling and, in so doing, boosts the production of IFN-I.
Finally, to investigate whether the antiviral function of IFI16 in IAV infection is dependent on RIG-I, A549 cells were transfected with the RIG-I- or MAVS-targeting siRNAs and then infected with IAV. Western blotting showed that the level of RIG-I or MAVS was significantly reduced by RIG-I-targeting siRNA-2835 (Extended Data Fig. 7i) or MAVS-targeting siRNA-571 transfection (Extended Data Fig. 6g). IAV infection could not effectively stimulate the production of IFN-β in RIG-I- or MAVS-silenced IFI16+/+ A549 cells (Extended Data Figs. 6h and 7j). Overexpression of IFI16 in RIG-I- or MAVS-silenced A549 cells could not induce the production of IFN-β during IAV infection (Extended Data Figs. 6l and 7k). RIG-I−/− HEK293 cells were then transfected with IFI16–Flag or Flag vectors and then infected with PR8 virus at 1 m.o.i. Overexpression of IFI16 failed to inhibit viral NP and M1 replication in PR8-virus-infected RIG-I−/− cells at 6 and 12 h.p.i. (Extended Data Fig. 7l), whereas exogenous expression of RIG-I in RIG-I−/− cells rescued the inhibitory effect of IFI16 on viral titres at 24 h.p.i. (Extended Data Fig. 7m). Taken together, our results indicate that the antiviral function of IFI16 is dependent on RIG-I.
Discussion
Host cells possess critical sensors that can discriminate viral and host nucleic acids. IFI16 has been demonstrated to be a sensor of viral DNA in innate immune signalling16,17. However, its role in sensing vRNA during infection, in particular influenza virus infection, is hitherto unknown. In this study, we demonstrated that IFI16 is a viral RBP and is induced during influenza virus infection. IFI16 inhibited influenza virus replication in human cell lines and in mice through an enhanced induction of IFN-I. IFI16 enhances the transcription of RIG-I, binds vRNAs and interacts with the RIG-I protein, thereby increasing the sensitivity of RIG-I signalling. These findings establish a critical role for IFI16 in antiviral innate immune responses to influenza virus and possibly other RNA viruses (a proposed signalling model is depicted in Extended Data Fig. 8).
We found that IFI16 was strongly induced by IAV, which is consistent with previous findings that IFI16 can be induced by RNA virus infection and be upregulated by the type I and type III IFNs as an ISG27,28,29. IFI16 is known to boost sensing of intracellular DNA and subsequent IFN-I induction21,24,26; it functions as a cytoplasmic immune-sensor-mediating pyroptotic death of tissue CD4+ T cells abortively infected with HIV-1 (ref. 22). Thus, the main role of IFI16 had been assumed to be a cytoplasmic sensor of viral DNA43. However, this is at odds with the findings that IFI16 is predominantly localized in the nucleus36. We found that IFI16 is induced in the nucleus and acts as a positive transcriptional regulator of RIG-I during IAV infection. These results confirm previous findings that IFI16 regulates RNA-virus-mediated IFN-I responses as a transcription factor27. We also found that IFI16 was induced in the cytoplasm and sensed and interacted with influenza vRNAs, which is consistent with recent findings that IFI16 directly binds to incoming chikungunya virus genome RNA and acts as a pattern recognition receptor30. Overexpression of IFI16 in cells significantly increased IFN-β expression and reduced viral titres, whereas knockdown or knockout of IFI16 had the opposite effects, which suggests that IFI16 is an important antiviral factor in IAV infection. Therefore, we demonstrated an important mechanism of IFI16 in RLR-mediated antiviral innate immune responses to influenza virus and furthered our understanding of the role of IFI16 in innate immunity.
The vRNA sensor RIG-I is thought to be the most important sensor of influenza virus infection9 in the recognition of cytoplasmic double-stranded RNA that leads to the transcriptional activation of IFN-I and downstream ISGs14. Thus, identification of positive regulators of RIG-I could be important in the control of virus infection. DNA-dependent activator of IFN-regulatory factor (DAI), a cytosolic DNA sensor, has already been found to recognize genomic RNA and regulate virus-induced cell-death pathways and thereby plays an important role in the pathogenesis of IAV infection44,45. Here, we demonstrated that IFI16 binds to vRNAs and enhances the RIG-I-mediated production of IFN-I during IAV infection. We further identified that IFI16 interacted with RIG-I via the PYRIN domain of IFI16. An interaction of IFI16 with RIG-I as a complex via siRNA was previously reported46. Notably, we found that IFI16 facilitates RIG-I binding to IAV vRNAs during infection and promotes virus-induced K63-linked polyubiquitination of RIG-I, thereby indicating that IFI16 positively contributes to RIG-I-dependent antiviral responses. It is currently not clear whether the filamentous structures of IFI16 are necessary for IFI16–vRNA and IFI16–RIG-I interactions or about the types of vRNA structures that are needed to interact with IFI16.
Collectively, our study showed that IFI16 induced by influenza virus infection positively regulates RIG-I signalling by enhancing its transcriptional expression through recruitment of RNA Pol II to the RIG-I promoter, and sensing of vRNA to promote virus-induced K63-linked polyubiquitination of RIG-I. This study highlights an important mechanism of IFI16 in RLR-mediated innate antiviral immune responses to IAV and possibly other RNA virus infections, and expands our understanding of the functions of the innate immune system in intracellular virus recognition, which could help the development of new strategies to modulate antiviral responses.
Methods
Ethics statement
All animal experiments were performed in accordance with institutional guidelines of China Agricultural University (CAU) (approval SKLAB-B-2010-003) and approved by the Beijing Association for Science and Technology of China (approval SYXK, Beijing, 2007–0023).
Cells
Madin–Darby canine kidney cells (MDCK), human embryonic kidney cells (HEK293) and human lung adenocarcinoma epithelial cells (A549) were maintained in-house. The THP-1 cell line was kindly provided by S. Zheng (China Agricultural University). IFI16−/− A549 and RIG-I−/− HEK293 cell lines were generously given by Y. Chen (Sun Yat-sen University) and W. Liu (Institute of Microbiology, Chinese Academy of Sciences), respectively. The IFNAR1−/− A549 cell line was generously given by Y. Zhu (Wuhan University), and the 2fTGH-ISRE cell line was kindly provided by F. You (Peking University)47. HEK293T-Gluc cells were kindly provided by S. Cen (Chinese Academy of Medical Sciences and Peking Union Medical School)48. Primary BMDMs were produced as previously described49. Cells were cultured in DMEM supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Gibco), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin.
Viruses
The influenza A/Puerto Rico/8/1934 (PR8; H1N1) strain was maintained in our laboratory. The PR8–GFP virus was generated by inserting the GFP CDS sequences at the carboxy terminal of NS1 as previously described50. The methods of cell culture, mice infection and virus titration in lung tissues were performed as previously described49.
Plasmid construction and transfection
IFI16, RIG-I, TBK1, TRIM25 and MAVS genes were amplified by PCR using PR8-virus-infected THP-1 cells at 8 h.p.i. Full-length and mutated IFI16 expression constructs were generated using PRK5 containing different tags or pCDNA3.1–GFP vectors by recombinase-mediated recombination. Full-length RIG-I expression vectors were created using PRK5–Flag or pCDNA3.1–mCherry vectors. Full promoter sequences or truncations of RIG-I were generated using a PGL3.0 luciferase reporter (Promega). Plasmid transfection experiments in HEK293 or A549 cells were performed using Lipofectamine 3000 reagent (Invitrogen). IFI16+/+ or IFI16−/− A549 cells were transfected with the indicated siRNAs using Lipofectamine RNAiMAX reagent (Invitrogen).
In vivo virus infection
p204−/− C57BL/6 mice were kindly provided by W. Tang (Shandong University). Sex- and age-matched WT C57BL/6 mice were purchased from Laboratory Animal Technology of Charles River, Beijing. Seven-week-old mice were intranasally inoculated with PR8 virus at a 50% tissue culture infectious dose (TCID50) of 50 in 50 μl of PBS. Body weight and survival were monitored daily after infection. Lung tissue lysates were generated by homogenizing snap-frozen lung tissues two times (20 s each time) in MEM medium and centrifuging the lung suspensions at 2,000 r.p.m. for 15 min. TCID50 assays were performed using MDCK cells and TCID50 values were calculated as previously described49. WT and p204−/− mice were intranasally innoculated with PR8 virus at a dose 100 TCID50. BALF was collected at 0 and 3 d.p.i. A BD CBA Mouse Inflammation kit (BD Biosciences, 552364) was used to quantitatively measure IL-6 and MCP-1 levels in BALF.
Antibodies and reagents
Anti-IFI16 (ab185812), anti-TBK1 (ab40676), anti-NP (ab104870), anti-ubiquitin (linkage-specific K63) (ab271929) and anti-RNA Pol II (ab238450) antibodies were from Abcam. Anti-IFI16 (sc-8023) and anti-TRIM25 (sc-166926) antibodies were from Santa Cruz. Rabbit anti-p65 (10745-1-AP), anti-HA (66006-2-Ig) and anti-RIG-I (20566-1-AP) antibodies were from Proteintech. Anti-IRF3 (YT2398) antibody was from ImmunoWay Biotechnology Company. Anti-phospho-TBK1 (Ser172; 5483); anti-phospho-IRF3 (Ser396; 4947), anti-IFI16 (14970S), anti-FLAG (8146), anti-His (9991) anti-Myc (2278) and anti-phospho-p65 (Ser536; 3033) antibodies were from Cell Signaling Technology. Poly(I:C) (TLRL-PIC) was from InvivoGen. Recombinant human IFN-γ (HY-P7025) and C-646 (HY-13823, 10 mg) was from MedChemExpress.
In vitro transcription and biotin-labelling RNA purification
Templates for T7 RNA transcription were linearized from H7N9 pSPT9 plasmids coding for individual RNA segments of H7N9 virus. T7 transcription reactions were carried out with T7 RNA polymerase in transcription buffer and biotin-dNTPs mix according to the manufacturer’s instructions (Promega)51. Following DNase I treatment, biotin-labelled vRNAs were extracted with phenol–chloroform, ethanol precipitated and purified with RNAeasy columns (Aidlab Biotechnologies) and analysed on denaturing agarose gels for correct size.
AP–MS
THP-1 cells were treated with phorbolmyristateacetate for 12 h, and infected with H7N9 virus (1 m.o.i.). After 12 h of infection, cells were lysed with lysis buffer and incubated with in vitro-transcribed 2 pM biotin-labelled vRNAs for 4 h followed by incubation with pre-washed Dynabeads M-280 streptavidin (Sigma) for 3 h at 4 °C. Beads were washed five times with NT2 buffer (50 mM Tris-HCl pH 7.0, 150 mM NaCl, 1 mM MgCl2 and 0.05% NP-40). Protein complex bound to the beads was then boiled with 20 μl PBS at 100 °C for 10 min. Pulled-down proteins were identified by liquid chromatography–MS.
Western blotting
Western blotting was performed as previously described49. Briefly, protein samples were mixed with loading buffer supplemented with 10% β-mercaptoethanol, heated at 95 °C for 5 min and separated on a 10% SDS–PAGE under reducing conditions. After electrophoresis, protein samples were electroblotted onto polyvinylidene difluoride membranes (Bio-Rad) and blocked for 2 h in Tris-buffered saline (10 mM Tris-HCl, pH 8.0, containing 150 mM NaCl) containing 5% (w/v) non-fat dry milk and 0.5‰ (v/v) Tween-20. The blots were incubated with the primary antibodies overnight at 4 °C. The next day, the blots were incubated with corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature (RT). HRP antibody binding was detected using a standard enhanced chemiluminescence (ECL) kit (Thermo Scientific).
RT–qPCR
Total RNA from virus-infected cells or lung tissues was extracted using an RNA isolation kit (Thermo Scientific). First-strand complementary DNA was synthesized from 1 μg of total RNA using a TransScript RT reagent kit (TransGen). Uni-12 primer was used for the detection of influenza vRNA, and oligo dT and random primers were used for detecting host and viral genes. Generated cDNA was subjected to qPCR in a 25-μl reaction volume using FastStart Universal SYBR Green master mix (Roche). Human β-actin (ACTB) and mouse GAPDH genes were amplified for normalization of the cDNA amount used in qPCR. Reactions were conducted in triplicate, and the data were analysed using the 2−ΔΔCt method. qPCR primers used in this study are provided in the Supplementary Information.
IFN-I bioassay
During PR8 virus infection, IFN-I released in the human cell culture medium was quantified in 2fTGH-ISRE cells as previously described52. In brief, 200 ml of cell culture supernatant was incubated with confluent 2fGTH-ISRE-Luci cells in a 24-well plate for 6 h. Cells were then lysed in passive lysis buffer and subjected to luciferase quantification (Promega). A serial dilution of recombinant human IFN-β (Invitrogen) was used as reference.
Luciferase reporter assays
HEK293 cells seeded on 24-well plates were co-transfected with 125 ng of the luciferase reporter plasmids and an equal amount of various expression plasmids or empty controls. Ten nanograms of pRL-TK plasmid was used as an internal control. At 24 h after transfection, Renilla luciferase and firefly luciferase expression levels were measured in the presence or absence of virus stimulation for 12 h using a Dual-Luciferase Reporter Assay kit (Promega) in a TD-20/20 Luminometer according to the manufacturers’ recommendations. Results were normalized to corresponding control reporter constructs.
Gaussia luciferase reporter assays were performed according to standard procedures described in detail elsewhere48. Briefly, HEK293T-Gluc cells were transfected with either IFI16–Flag or Flag control plasmids followed by infection with PR8 virus (1 m.o.i.) for 24 h, and viral infectivity was evaluated by measuring Gluc activity. After a further 24 h of incubation, cell supernatants were collected and measured for Gluc activity. Dimethylsulfoxide was used as negative controls. The inhibition rate of the tested compounds was calculated with the following equation, where RLU indicates relative light unit:
Histology
WT and p204−/− mice were euthanized at the indicated time points after PR8 virus infection. Lung tissues were collected and fixed with 4% formaldehyde, followed by paraffin embedding. For histopathological analysis, 5- to 7-μm sections were sectioned longitudinally through the left and right lung and stained using a standard haematoxylin and eosin (H&E) protocol.
Immunohistochemistry
Lung sections were deparaffinized with xylene and rehydrated with ethanol gradations and water. Endogenous peroxidase activity was blocked using 3% hydrogen peroxide in methanol. PBS containing 0.05% Tween-20 was used to wash lung tissue sections between steps. Lung sections were incubated with the primary anti-NP antibody (ab20343) at 1:100 dilution or with the isotype control at the same concentration at 4 °C overnight in a humidified chamber. Sections were subsequently incubated with the HRP-conjugated secondary antibody for 60 min at RT. Immunodetection was performed using a Vector Elite ABC kit (Vectastain, Vector).
Co-IP
For the co-IP assays, A549 cells were infected with PR8 virus (5 m.o.i.). After infection, cell samples were collected and lysed in 800 μl of IP lysis buffer (Thermo Scientific) containing protease and phosphatase inhibitors. A portion of each whole cell lysate sample was kept to confirm protein expression levels, and 500 μg of cell lysate was used for the co-IP assays. Lysates were incubated with anti-IFI16, anti-RIG-I or IgG antibodies overnight at 4 °C under constant rotation, and then 40 μl of protein A/G beads (Santa Cruz) were added and incubated for 2 h at 4 °C under gentle rotation. The beads were then washed four times with cold lysis buffer and analysed by SDS–PAGE and western blotting.
RIP–qPCR
Two 10-cm2 dishes (107 cells per dish) of A549 cells were infected with PR8 virus for 12 h. Cells were lysed with RIP lysis buffer (50 mM HEPES, 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5% NP40, 0.5 mM dithiothreitol (DTT), 1× protease inhibitor cocktail and 25 U RNasin) for 30 min at 4 °C. Cell lysates were centrifuged at 12,000 r.p.m. for 15 min at 4 °C and the supernatants were subjected to RIP. A 50 μl aliquot of cell supernatant was saved as input, and the remaining samples were each incubated with 5 μg anti-IFI16 antibody or IgG antibody and 40 μl protein A/G beads for 10 h at 4 °C under gentle shaking. After IP, the beads were pelleted and washed four times with RIP wash buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM MgCl2 and 0.05% NP40), resuspended in 250 ml of DNase digestion buffer (40 mM Tris pH 8.0, 10 mM MgSO4 and 1 mM CaCl2) and treated with 25 U RNasin (Promega) and 2 U DNase I (NEB) at 37 °C for 20 min. Beads were then washed and resuspended in 100 μl RIP wash buffer, 10% of each sample was removed for immunoblot analysis. Samples were treated with 4 U proteinase K at 55 °C for 30 min. The input and immunoprecipitated RNAs were isolated by 1 ml of TRIzol reagent (Sigma), and vRNAs were analysed by RT–qPCR or RNA sequencing.
ChIP–qPCR
Approximately 107 treated cells were cross-linked with 1% formaldehyde at RT for 10 min, and the crosslinking was quenched with 0.125 M glycine for 5 min. Cells were then collected by centrifugation, lysed with SDS lysis buffer containing protease inhibitor cocktail and sonicated to shear the DNA. The sonicated DNA–protein complexes were incubated with anti-IFI16 (Cell Signaling Technology, D8B5T), anti-RNA Pol II (Abcam, ab238450) or control IgG (Beyotime, A7016) antibodies. The immunocomplexes were collected using Protein G Dynabeads (10004d, Invitrogen). The Dynabeads were washed once with wash buffer A (20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 2 mM EDTA, 0.1% SDS and 1% Triton X-100), once with wash buffer B (10 mM Tris-HCl (pH 8.0), 250 mM LiCl, 1 mM EDTA and 1%NP-40) and three times with wash buffer C (1 mM EDTA, 10 mM Tris-HCl, pH 8.0). The beads were eluted with 100 ml elution buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA and 1% SDS) followed by incubation at 65 °C overnight to reverse crosslinking. The next day, the DNA was purified with a QIAquick PCR purification kit (Magen, D211102) and analysed using FastStart Universal SYBR Green master mix (Roche).
Confocal microscopy
A549 or HEK293 cells on coverslips were washed twice with pre-warmed PBS and fixed with 4% paraformaldehyde for 15 min at RT. Cells were subsequently permeabilized with immunostaining permeabilization buffer containing Triton X-100 (Beyotime) for 10 min and blocked in QuickBlock blocking buffer for 20 min at RT. Fixed cells were incubated with indicated antibodies diluted in immunostaining primary antibody dilution buffer at 4 °C overnight. Coverslips were then washed three times with PBS and incubated with Alexa Fluor 488-conjugated secondary antibodies or Alexa Fluor 555-conjugated secondary antibodies for 1 h at 37 °C. Coverslips were finally washed three times and mounted onto microscope slides with 4,6-diamidino-2-phenylindole (DAPI) staining solution (Beyotime) for 8 min and examined by confocal microscopy. Immunostained cells were visualized using a Nikon super-resolution laser scanning confocal microscope under a ×100 oil objective and analysed using the Imaris 9.2 platform.
In situ PLA microscopy
A DuoLink PLA kit (DUO92105-1KT, Sigma) was used to test protein–protein interactions as described in the protocol. WT or IFI16−/− A549 cells were infected with PR8 virus at 1 m.o.i. for 12 h, fixed and permeabilized as described in the “Confocal microscopy” section and blocked with DuoLink blocking buffer for 30 min at 37 °C. Cells were incubated with corresponding primary antibodies diluted in DuoLink dilution buffer. After washing, cells were incubated with species-specific PLA probes (Plus and Minus) for 1 h at 37 °C under hybridization conditions and in the presence of two additional oligonucleotides to facilitate hybridization of PLA probes if they were in close proximity (<40 nm). Ligase was then added and incubated for 30 min at 37 °C to join hybridized oligonucleotides. Amplification polymerase was added to generate a concatemeric product extending from the oligonucleotide arm of the PLA probe. Finally, a detection solution consisting of fluorescently labelled oligonucleotides was added, and the labelled oligonucleotides were hybridized to the concatemeric products. Nuclei was stained with Duolink in situ mounting medium containing DAPI.
Ubiquitination assay
HEK293 cells were transfected with plasmids encoding RIG-I–Flag, TRIM25-His with or without coexpression of IFI16–Myc or Myc empty vectors, and HA-ubiquitin (WT) or HA-ubiquitin mutants (K48 or K63). After 24 h of transfection, cells were collected and lysed in RIPA buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 0.1% SDS and 1 mM EDTA) containing protease inhibitor cocktail and 10 µM deubiquitinase inhibitor N-ethylmaleimide (Sigma). The cell extracts were immunoprecipitated with anti-Flag antibody overnight at 4 °C and then beads were added to the samples for 1–1.5 h at 4 °C. The beads were washed three times with RIPA buffer and analysed by immunoblotting with anti-HA antibody.
RNA FISH
A549 cells were grown in 24-well slide chambers and infected with PR8 at a m.o.i. of 1. At 12 h.p.i., cells were fixed for 15 min in 4% paraformaldehyde, then permeabilized and dehydrated by sequential 3-min incubations as follows: once with 70% ethanol, once with 85% ethanol and three times with 100% ethanol. Cells were then hybridized with the Alexa Fluor 488-conjugated NP RNA target probes (NP-probes, GenePharma) of PR8 virus in hybridization buffer for 10 min at 75 °C. Cells were further incubated for 12–16 h at 37 °C. Finally, cells were stained with anti-IFI16 and secondary antibodies, and nuclei were stained with DAPI as previously described.
MST
HEK293 cells were separately transfected with the HINa–GFP, HINb–GFP and PYRIN–GFP expression vectors. After 24 h of transfection, cell lysates were collected and incubated with twofold serial dilutions of indicated vRNAs in MST-optimized buffer at a constant concentration (20–100 nM). Equal volumes of binding reactions were mixed by pipetting and incubated for 15 min at RT. Mixtures were enclosed in standard-treated glass capillaries and loaded into the instrument (Monolith NT.115, NanoTemper). To identify whether IFI16 directly binds to the IAV RNAs, the transcribed RNAs were labelled with fluorescein RNA labelling mix (11685619910, Sigma) and purified as previously described53. Purified GST–IFI16 proteins (Abcam, ab158724) were incubated with different amounts of IAV full-length fluorescein-labelled RNAs, followed by MST assays. For all the measurements, 200–1,000 counts were obtained for the fluorescence intensity. Dissociation constant (Kd) values were determined using the NanoTemper analysis tool.
RNA pull-down assay
HEK293 cells were transfected with Flag-tagged RIG-I, IFI16, TBK1 or MAVS vectors. After 24 h of transfection, cells were collected and lysed with lysis buffer (50 mM Tris-HCl pH 7.0, 150 mM NaCl, 1 mM MgCl2 and 0.05% NP-40). Cell lysates were mixed with transcribed biotin-labelled viral NP RNA for 4 h at 4 °C and incubated with pre-washed Dynabeads M-280 Streptavidin (Sigma) for another 3 h at 4 °C. The protein samples bound to the beads were boiled and analysed by SDS–PAGE and western blotting.
DNA pull-down assay
IFI16 proteins were purified from IFI16–Flag-overexpressing HEK293 cells by immunoprecipitation using M2 beads (Sigma). Biotinylated mutant DNA probes were synthesized using an EMSA Probe Biotin Labelling kit (Beyotime, GS008) and were annealed and incubated with the purified Flag-tagged IFI16 proteins for 30 min in binding buffer (10 mM Tris, 1 mM KCl, 1%NP-40, 1 mM EDTA and 5% glycerol) at RT. Then, 40 μl prewashed Dynabeads M-280 Streptavidin (Sigma) was added for incubation at 4 °C for 1 h. The mutant probe-binding proteins were eluted by boiling and analysed by immunoblotting.
Statistical analysis
For all the bar graphs, data are shown as the mean ± s.e.m. All statistical analyses were performed using GraphPad Prism software v.7.00 (GraphPad Software). The Kaplan–Meier method was employed for survival analysis. Differences in means were considered statistically significant at P < 0.05. And significance levels are as follow: *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The MS proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE54 partner repository (https://www.ebi.ac.uk/pride/) with the dataset identifiers PXD020723 and PXD020723. The accession numbers for the RNA sequencing data are GSE157609 and GSE158122. Source data are provided with this paper.
References
Collins, P. J. et al. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453, 1258–1261 (2008).
Wang, M. Z., Tai, C. Y. & Mendel, D. B. Mechanism by which mutations at His274 alter sensitivity of influenza A virus N1 neuraminidase to oseltamivir carboxylate and zanamivir. Antimicrob. Agents Chemother. 46, 3809–3816 (2002).
Deyde, V. M. et al. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 196, 249–257 (2007).
Furuse, Y., Suzuki, A. & Oshitani, H. Large-scale sequence analysis of M gene of influenza A viruses from different species: mechanisms for emergence and spread of amantadine resistance. Antimicrob. Agents Chemother. 53, 4457–4463 (2009).
Eisfeld, A. J., Neumann, G. & Kawaoka, Y. At the centre: influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 13, 28–41 (2015).
Blasius, A. L. & Beutler, B. Intracellular Toll-like receptors. Immunity 32, 305–315 (2010).
Takeuchi, O. & Akira, S. Innate immunity to virus infection. Immunol. Rev. 227, 75–86 (2009).
Pang, I. K. & Iwasaki, A. Inflammasomes as mediators of immunity against influenza virus. Trends Immunol. 32, 34–41 (2011).
Kato, H. et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006).
Pichlmair, A. et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314, 997–1001 (2006).
Gack, M. U. et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007).
Oshiumi, H. et al. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8, 496–509 (2010).
Jiang, X. et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36, 959–973 (2012).
Loo, Y. M. & Gale, M. Jr. Immune signaling by RIG-I-like receptors. Immunity 34, 680–692 (2011).
Dawson, M. J. & Trapani, J. A. The interferon-inducible autoantigen, IFI 16: localization to the nucleolus and identification of a DNA-binding domain. Biochem. Biophys. Res. Commun. 214, 152–162 (1995).
Orzalli, M. H., Conwell, S. E., Berrios, C., DeCaprio, J. A. & Knipe, D. M. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc. Natl Acad. Sci. USA 110, E4492–E4501 (2013).
Johnson, K. E. et al. IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog. 10, e1004503 (2014).
Almine, J. F. et al. IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Commun. 8, 14392 (2017).
Goubau, D., Deddouche, S. & Reis e Sousa, C. Cytosolic sensing of viruses. Immunity 38, 855–869 (2013).
Jonsson, K. L. et al. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 8, 14391 (2017).
Kerur, N. et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9, 363–375 (2011).
Monroe, K. M. et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343, 428–432 (2014).
Orzalli, M. H. et al. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl Acad. Sci. USA 112, E1773–E1781 (2015).
Unterholzner, L. et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 (2010).
Ansari, M. A. et al. Herpesvirus genome recognition induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN-beta responses. PLoS Pathog. 11, e1005019 (2015).
Dunphy, G. et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-ΚB signaling after nuclear DNA damage. Mol. Cell 71, 745–760.e5 (2018).
Thompson, M. R. et al. Interferon γ-inducible protein (IFI) 16 transcriptionally regulates type I interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J. Biol. Chem. 289, 23568–23581 (2014).
Cao, L. et al. P200 family protein IFI204 negatively regulates type I interferon responses by targeting IRF7 in nucleus. PLoS Pathog. 15, e1008079 (2019).
Chang, X. et al. IFI16 inhibits porcine reproductive and respiratory syndrome virus 2 replication in a MAVS-dependent manner in MARC-145 cells. Viruses https://doi.org/10.3390/v11121160 (2019).
Kim, B. et al. Discovery of widespread host protein interactions with the pre-replicated genome of CHIKV using VIR-CLASP. Mol. Cell 78, 624–640 (2020).
Monzon-Casanova, E. et al. The RNA-binding protein PTBP1 is necessary for B cell selection in germinal centers. Nat. Immunol. 19, 267–278 (2018).
Grellscheid, S. N. et al. Molecular design of a splicing switch responsive to the RNA binding protein Tra2β. Nucleic Acids Res. 39, 8092–8104 (2011).
Li, H. et al. RNA helicase DDX5 inhibits reprogramming to pluripotency by miRNA-based repression of RYBP and its PRC1-dependent and -independent functions. Cell Stem Cell 20, 462–477.e6 (2017).
Herdy, B. et al. Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res. 46, 11592–11604 (2018).
Di Giammartino, D. C. et al. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3′ UTRs. Genes Dev. 28, 2248–2260 (2014).
Li, T., Diner, B. A., Chen, J. & Cristea, I. M. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl Acad. Sci. USA 109, 10558–10563 (2012).
Baum, A., Sachidanandam, R. & Garcia-Sastre, A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl Acad. Sci. USA 107, 16303–16308 (2010).
Jakobsen, M. R. & Paludan, S. R. IFI16: at the interphase between innate DNA sensing and genome regulation. Cytokine Growth Factor Rev. 25, 649–655 (2014).
Li, D. et al. STING-mediated IFI16 degradation negatively controls type I interferon production. Cell Rep. 29, 1249–1260 (2019).
Merkl, P. E. & Knipe, D. M. Role for a filamentous nuclear assembly of IFI16, DNA, and host factors in restriction of herpesviral infection. mBio https://doi.org/10.1128/mBio.02621-18 (2019).
Antiochos, B., Matyszewski, M., Sohn, J., Casciola-Rosen, L. & Rosen, A. IFI16 filament formation in salivary epithelial cells shapes the anti-IFI16 immune response in Sjogren’s syndrome. JCI Insight https://doi.org/10.1172/jci.insight.120179 (2018).
Morrone, S. R. et al. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc. Natl Acad. Sci. USA 111, E62–E71 (2014).
Hotter, D. et al. IFI16 targets the transcription factor Sp1 to suppress HIV-1 transcription and latency reactivation. Cell Host Microbe 25, 858–872 (2019).
Thapa, R. J. et al. DAI senses influenza a virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe 20, 674–681 (2016).
Kuriakose, T. et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol. https://doi.org/10.1126/sciimmunol.aag2045 (2016).
Sui, H., Zhou, M., Chen, Q., Lane, H. C. & Imamichi, T. siRNA enhances DNA-mediated interferon lambda-1 response through crosstalk between RIG-I and IFI16 signalling pathway. Nucleic Acids Res. 42, 583–598 (2014).
Cao, L. et al. The nuclear matrix protein SAFA surveils viral RNA and facilitates immunity by activating antiviral enhancers and super-enhancers. Cell Host Microbe 26, 369–384 (2019).
Gao, Q. et al. A cell-based high-throughput approach to identify inhibitors of influenza A virus. Acta Pharm. Sin. B 4, 301–306 (2014).
Wei, F. et al. Induction of PGRN by influenza virus inhibits the antiviral immune responses through downregulation of type I interferons signaling. PLoS Pathog. 15, e1008062 (2019).
Manicassamy, B. et al. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl Acad. Sci. USA 107, 11531–11536 (2010).
Gavazzi, C. et al. An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: comparison with a human H3N2 virus. Nucleic Acids Res. 41, 1241–1254 (2013).
You, F. et al. ELF4 is critical for induction of type I interferon and the host antiviral response. Nat. Immunol. 14, 1237–1246 (2013).
Zhang, Y. et al. Identifying local and descending inputs for primary sensory neurons. J. Clin. Invest. 125, 3782–3794 (2015).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Acknowledgements
We thank F. You (Peking University), W. Liu (Institute of Microbiology, Chinese Academy of Sciences), Y. Zhu (Wuhan University) and Y. Chen (Wuhan University) for kindly providing cell lines, and W. Tang (Shandong University) for generously gifting the p204−/− mice. This work was supported by the National Key Research and Development Program of China (2016YFD0500204 to J.L.) and the National Natural Science Foundation of China (81960297 to F.W.).
Author information
Authors and Affiliations
Contributions
Z.J. and F.W. performed and analysed most of the experiments. Y.Z., T.W. and W.G. performed the AP–MS experiments. S.Y., H.S., J.P., Y.S., M.W. and Q.T. generated biochemical reagents. C.G. and K.-C.C. guided and analysed the data. F.W. and J.L. conceived and supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Microbiology thanks Peter Staeheli, Aartjan te Velthuis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 IFI16 induction by IAV is dependent on viral replication.
a–c, IFI16 expression in a, THP-1, b, A549, and c, HEK293 was quantified by RT-qPCR. (d) p204 expression in lung tissues from PR8 virus-infected WT mice was determine. e, IFI16 expression in 1.0 MOI of PR8 virus-infected THP-1 cells was detected. f, IFI16 mRNA expression in 2 MOI of UV-inactivated and live PR8 virus-infected A549 cells was determined. g, IFI16 protein expression in 2 MOI of UV-inactivated PR8 virus-infected A549 cells was determined. h, IFI16 expression in the nuclear and cytoplasmic fractions of PR8 virus-infected A549 cells were determined. β-actin and H3 were used as purity markers for cytoplasmic and nuclear fractions, respectively. i, IFI16 expression in A549 cells transfected with poly(I:C) for 18 h was determined by Western blotting. j, IFI16 expression in A549 cells treated with IFN-γ for 18 h was determined. (k) Intracellular localization of IFI16 in A549 cells treated with poly(I:C) or IFN-γ for 12 h was determined. Scale bars, 45 μm and 5 μm (enlarged). l, A549 cells were infected with PR8 virus at 0, 6, 12 and 24 hpi. Cell lysates were then immunoprecipitated with anti-acetylated lysine. Bound proteins were analyzed by immunoblots with anti-IFI16 antibody. m, A549 cells were pre-incubated with C-646 for 2 h, then infected with PR8 virus for 1 h, washed and incubated in complete medium with or without C-646. IFI16 expression and viral NP protein in the nuclear and cytoplasmic fractions of PR8 virus-infected A549 cells at indicated time points were determined by Western blotting. β-Actin and H3 were used as purity markers for cytoplasmic and nuclear fractions, respectively. (a–d) and f, Data presented as means ± SD from three independent experiments. (e) and g–m, Data are representative of three independent experiments. Statistical significance in (a) to (d) and (f) was determined by unpaired two-tailed Student’s t-test.
Extended Data Fig. 2 IFI16 inhibits IAV viral replication.
a, A549 cells were transfected with IFI16-Flag plasmids or empty control for 24 h and then infected with PR8 virus at 2.0 MOI. mRNA and vRNA expression of NP and M genes at 6, 12 and 18 hpi were determined by RT-qPCR. Data are presented as means ± SEMs from three independent experiments. b, HEK293 cells were transfected with IFI16-Flag plasmids or empty control for 24 h and then infected with PR8 virus at 2.0 MOI. NP protein expression at 0, 6, 12 and 18 hpi was determined by Western blotting. β-Actin detection was used as loading control. (c) and (d), A549 cells were transfected with IFI16-targeting siRNA and negative control (NC) siRNA for 24 h, followed by infection with PR8 virus (MOI = 1). c, NP and M1 protein expression were determined by Western blotting at 0, 12, 18 and 24 hpi. β-Actin detection was used as loading control. d, Viral titers were determined by TCID50 assay at the 24 hpi. Data presented as means ± SD from three independent experiments. (b) and (c), Data are representative of three independent experiments. Statistical significance in (a) and (d) was determined by unpaired two-tailed Student’s t-test. ns = non-significant.
Extended Data Fig. 3 IFI16 enhances the activation of IFN-I pathway during IAV infection.
a to c, Serum-starved A549cells were transfected with IFI16-encoding plasmids or negative control, and then infected with 1.0 MOI of PR8 virus. Expression of IFN-β (a), ISG15 (b), IL-6 (c) at the indicated time points was determined by RT-qPCR. d to f, mRNA expression of IFN-α4 (d), IRF7 (e) and CXCL5 (f) in PR8 virus-infected IFI16+/+ and IFI16−/− A549 cells at 0, 6, and 12 hpi was determined by RT-qPCR. (g) Immunoblot analysis of RIG-I-triggered downstream signaling pathway in IFI16+/+ and IFI16−/− A549 cells after stimulation with 5′ppp-RNA for the indicated duration. h, IFI16+/+ and IFI16−/− A549 cells transfected with IFI16-Flag expression vectors or empty control for 24 h and then infected with PR8 virus at 1.0 MOI. RIG-I-triggered downstream signaling pathway at 0, 4, 8 and 12 hpi was assessed with indicated antibodies. a to f, Data presented as means ± SD from three independent experiments. g to h, Data are representative of three independent experiments. Statistical significance in (a) to (f) was determined by unpaired two-tailed Student’s t-test. ns = non-significant.
Extended Data Fig. 4 IFI16 binds RIG-I promoter and enhances RIG-I transcription.
a, RIG-I expression in A549 cells transfected with IFI16-Flag vectors was examined. b, HEK293 cells co-transfected with indicated plasmids for 24 h were treated with cycloheximide (CHX) for 12 and 24 h. Expression of RIG-I and IFI16 in cell lysates was detected. c, RIG-I expression in ifnar1−/−A549 cells transfected with IFI16-Flag vectors was examined. d, RIG-I mRNA expression in ifnar1−/− A549 cells transfected with IFI16-Flag plasmids for 24 h was determined. e, RIG-I expression in A549 cells transfected with IFI16-Flag plasmids for 24 h was determined. f, RIG-I expression in lung tissues from virus-infected WT and p204−/− mice (n = 3) was determined. g, Schematic diagram of the biotinylated probe sequences from the promoter of RIG-I gene and corresponding mutants. h, IB analysis of the binding ability between wild type (p1 and p2) and mutated (p2-mut1 to p2-mut5) probes, and IFI16 proteins. i, IFI16+/+ and IFI16−/− A549 cells were infected with PR8 virus for 12 h, followed by ChIP assay. j, Schematic diagram of full-length IFI16 vectors and truncated mutants. k, A549 cells were transfected with Flag-tagged full-length IFI16 vectors and truncated mutant plasmids. After 24 h transfection, nuclear extracts were incubated with non-biotinylated or biotinylated promoter sequence of RIG-I for 4 h. Nuclear extracts were examined for IFI16 and truncated mutant expression by Western blotting. l, A549 cells after Flag-tagged IFI16 vectors and truncated mutants transfection and PR8 infection was determined. d to f and i, Data are representative of three independent experiments (mean ± SD). (a), (b), (c), (h), (k), and (l), Data are representative of three independent experiments. Statistical significance in (d) to (f) and (i) was determined by unpaired two-tailed Student’s t-test. ns = non-significant.
Extended Data Fig. 5 IFI16 directly binds viral RNA during infection.
a, HEK293 cells were transfected with indicated vectors for 24 h. Cell lysates were incubated with biotin-labeled viral NP RNA and immunoprecipitated with streptavidin beads. Bound proteins were analyzed by immunoblots with anti-Flag antibody. b, HEK293 cells were transfected with HINa-GFP, HINb-GFP, and PYRIN-GFP expression vectors for 24 h. Cell lysates were incubated with influenza NS vRNA. Binding affinity between indicated proteins and NS vRNA was determined by MST assays. c, Purified GST-IFI16 proteins were incubated with fluorescein labeled influenza HA, NP, PA and PB2 vRNAs. Binding affinity was determined by MST assays. d, PCR detection of NP vRNA in eluted RNA from RIG-I-Flag, IFI16-Flag and indicated Flag-tagged IFI16 truncated constructs. The data represent means ± SD. (n = 3 independent experiments). e, PCR detection of NA vRNAs in eluted RNA from RIG-I-Flag, IFI16-Flag and indicated Flag-tagged IFI16 truncations. Data were normalized to vRNA from RIG-I-Flag immunoprecipitates. The data represent means ± SD. (n = 3 independent experiments). f, Confocal microscopy detecting the co-localization of endogenous IFI16 and RIG-I in PR8 virus-infected A549 cells at 0, 6 and 18 hpi. Nuclei were stained with DAPI (left). Scale bars, 10 μm. Quantification of the co-localization of IFI16 and RIG-I in cells (right). Means ±SD from 3 biological samples. g, Confocal microscopy of HEK293 cells transfected with RIG-I-mCherry plasmids, together with expression vectors for IFI16-GFP or GFP-tagged mutants. DAPI serves as a marker for nuclei (left). Scale bars, 20 μm. Quantification of the co-localization of IFI16 or mutants and RIG-I in cells (right). Means ±SD from 3 biological samples. h, Co-IP analysis of the interaction between Myc-tagged RIG-I and Flag-tagged full-length or mutant IFI16 in HEK293 cells. (i) Three-dimensional confocal microscopy of HEK293 cells co-transfected with plasmids encoding PYRIN-GFP and RIG-I-mCherry. DAPI serves as a marker for nuclei. All data are representative of three independent experiments. Scale bars, 5 μm.
Extended Data Fig. 6 RIG-I is involved in IFI16-mediated antiviral response in IAV infection.
a, IFI16+/+ or IFI16−/− A549 cells were infected with PR8 virus (MOI = 1) and then analyzed by PLA. The right panels are enlarged. Red point represents TRIM25 plus RIG-I complexes. Scale bars, 23.8 μm (left) and 7.45 μm (enlarged). b, IFI16+/+ or IFI16−/− A549 cells were infected with PR8 virus (MOI = 1) and then analyzed by PLA. The right panels are enlarged. Red point indicates K63 Ub plus RIG-I complexes. Scale bars, 40.3 μm (left) and 14.3 μm (enlarged). c, RIP-EMSA analysis of the binding of biotinylated PA vRNA to purified Flag-RIG-I protein by adding purified GST-IFI16 at dose of 50 and 200 ng. d, RNA pull-down analysis of the binding of biotinylated PA vRNA to purified Flag-RIG-I protein by adding purified GST-IFI16 at dose of 30, 60, and 120 ng. e, A549 cells were transfected with Flag-tagged IFI16, its truncated forms and control. RNA eluted from RIG-I immunoprecipitates were detected by RT-qPCR with specific primers of PA gene. f, A549 cells were transfected with Flag-tagged IFI16 and its truncated forms for 24 h and then infected with PR8 virus. Production of IFN-β in supernatants at 18 hpi was determined. g, Knockdown effect of MAVS-targeting siRNAs in A549 cells. h, IFI16+/+ or IFI16−/− A549 cells were transfected with control or MAVS-targeting siRNA #571 and after 24 h infected with PR8 virus. Production of IFN-β in supernatants at 18 hpi was determined (i) IFI16+/+ A549 cells were transfected with negative control or MAVS-targeting siRNA #571, and 24 h later, cells were transfected with Flag-IFI16 plasmids or control; 24 h later cells were infected with PR8 virus. Production of IFN-β in supernatants on 18 hpi were quantified. (e), (f), (h), and (i), Data are representative of three independent experiments (mean ± SD). (a) to (d) and (g), All data are representative of three independent experiments. Statistical significance in (e) to (f) and (i) was determined by unpaired two-tailed Student’s t-test. ns = non-significant.
Extended Data Fig. 7 IFI16 promotes RIG-I signaling.
a, The interaction between TRIM25, RIG-I, and K63-linked ubiquitination of RIG-I in PR8 virus-infected IFI16+/+ and IFI16−/− A549 cells. b, Co-IP analysis of RIG-I ubiquitination in HEK293 cells transfected with indicated plasmids. c, Cell lysates were prepared after indicated plasmids transfection and PR8 infection and then immunoprecipitated with control IgG or anti-Flag. Bound-RNA was extracted for qPCR analysis. d, IFI16+/+ and IFI16−/− A549 cells were infected with PR8 virus for 12 h. Cell lysates were then immunoprecipitated with IgG or anti-RIG-I. Bound-RNA was extracted for analysis. e, Schematic experimental procedure used in (f). f, RNA co-purified with RIG-I from IAV-infected IFI16+/+ and IFI16−/− A549 cells were transfected into A549 cells, and IFN-β was determined. g, Luciferase activity of HEK293 cells after transfection with IFI16 vectors or control and then infection with PR8 virus. h, Luciferase activity of HEK293 cells transfected with an IFN-β reporter plasmids, RIG-I vectors, and control or IFI16 vectors. i, Knockdown effect of RIG-I-targeting siRNAs in A549 cells. j, IFN-β in IFI16+/+ or IFI16−/− A549 cells after transfection with siRNA #2835 or control and infection with PR8 virus. k, IFN-β in IFI16+/+ cells after transfection with siRNA #2835 or control, IFI16-Flag or control plasmids, and infection with PR8 virus. l, NP protein levels in RIG-I−/− HEK293 cells after transfection with IFI16-Flag vectors or control and infection with PR8 virus were determined. m, RIG-I−/− HEK293 cells were transfected with indicated plasmids for 24 h and then infected with PR8 virus at 1.0 MOI. Viral titers were determined. (a) to (d), (i), and (l), Data are representative of three independent experiments. (c) to (d), (f) to (h), (j), (k) and (m), Data presented as means ± SD from three independent experiments, and the significance of the results was assessed using a parametric paired t-test (Student’s two-tailed t-test). ns = non-significant.
Extended Data Fig. 8 Schematic model to show IFI16 enhances RIG-I signaling in influenza virus infection.
Briefly, influenza virus infection upregulates IFI16 expression; IFI16 protein, in turn, enhances transcription of RIG-I. In addition, IFI16 protein, via its HINa domain, directly senses viral RNA and, via its PYRIN domain, interacts with RIG-I receptor to promote antiviral RIG-I signaling.
Supplementary information
Supplementary Information
The qPCR or PCR primers sequence used in this study.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Jiang, Z., Wei, F., Zhang, Y. et al. IFI16 directly senses viral RNA and enhances RIG-I transcription and activation to restrict influenza virus infection. Nat Microbiol 6, 932–945 (2021). https://doi.org/10.1038/s41564-021-00907-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-021-00907-x
This article is cited by
-
Interferon-α stimulates DExH-box helicase 58 to prevent hepatocyte ferroptosis
Military Medical Research (2024)
-
UiO-66 nanoparticles combat influenza A virus in mice by activating the RIG-I-like receptor signaling pathway
Journal of Nanobiotechnology (2024)
-
The role of inflammasomes in human diseases and their potential as therapeutic targets
Signal Transduction and Targeted Therapy (2024)
-
Evolution of the Major Components of Innate Immunity in Animals
Journal of Molecular Evolution (2024)
-
Expression of down-regulated ERV LTR elements associates with immune activation in human small-cell lung cancers
Mobile DNA (2023)