Abstract
We are now in a time of readily available brain imaging data. Not only are researchers now sharing data more than ever before, but additionally large-scale data collecting initiatives are underway with the vision that many future researchers will use the data for secondary analyses. Here I provide an overview of available datasets and some example use cases. Example use cases include examining individual differences, more robust findings, reproducibility–both in public input data and availability as a replication sample, and methods development. I further discuss a variety of considerations associated with using existing data and the opportunities associated with large datasets. Suggestions for further readings on general neuroimaging and topic-specific discussions are also provided.
Similar content being viewed by others
Introduction
It is a great time to be studying human brain development, aging, or differences between healthy individuals and variety of neurological patient conditions–massive amounts of already acquired and openly available MRI data exist. As long as you’re satisfied with existing data collection protocols (e.g., not developing a new MR sequence or cognitive task), the data you are looking for to test a novel measure of brain structure, connectivity, or task-related activation may be only a few clicks away. Data sharing has countless benefits, allowing for the ready assessment of new research questions, enhancing reproducability, providing initial ‘pilot’ data for new methods development, and reducing the costs associated with doing neuroimaging research (Mar et al. 2013; Poldrack and Gorgolewski 2014; Madan 2017; Milham et al. 2018). While there are some considerations needed related to over-fitting to specific datasets (Madan 2017), otherwise referred to as ‘dataset decay’ (Thompson et al. 2020), there is much we can learn from these existing datasets before we must go out and acquire new ones.
The availability of data sharing has greatly increased over the last few years, in no small part due to the development of the ‘FAIR guiding principles for scientific data management and stewardship’ (Wilkinson et al. 2016): Findability, Accessibility, Interoperability, and Reuse of digital assets. Adherence to the FAIR guidelines is further facilitated by consistent file organisation standards (i.e., Brain Imaging Data Structure; BIDS) (Gorgolewski et al. 2016). Other standards and guidelines are also advancing the methodological rigor of the field, such as the Committee on Best Practices in Data Analysis and Sharing (COBIDAS) MRI (Nichols et al. 2017), among other best-practice recommendations (Eglen et al. 2017; Shenkin et al. 2017). Typical MRI studies can be readily shared using platforms including OpenNeuro (Poldrack and Gorgolewski 2017), allowing for further analyses of the data by other research groups, as well as assessments of analysis reproducability, though large-scale projects may require more dedicated infrastructure (discussed later).
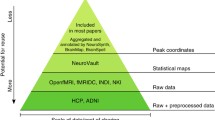
Here I will focus on the availability of large-scale neuroimaging datasets that help us move beyond the statistical power issues that are still typical within the field (Button et al. 2013; Zuo et al. 2019) and more towards furthering our understanding of the brain. This shift towards large-scale datasets can also be important for individual analyses, as these large datasets provide a more meaningful opportunity to shift from group averaging to comparing the statistics of individual participants (Dubois and Adolphs 2016), bolstered by multimodal acquisitions and highly sampled individuals, providing richer insights into individual brains and their relative differences. Naselaris et al. (2021) provides an insightful discussion for considering trade-offs between sampling more individuals as compared to more experimental data from a few individuals (e.g., considering a fixed amount of total scan time), as summarised in Fig. 1.
Trade-offs between number of participants and amount of data per participant. Note that some datasets have increased in size since the generation of this figure (e.g., IBC has more data per participant now); some datasets are not featured in the current review, e.g., VIM-1. Reprinted from Naselaris et al. (2021)
The magnitude of considerations needed when designing a large-scale dataset cannot be understated. For instance, a myriad of topics related to the design of the Human Connectome Project (HCP) were discussed in a special issue of NeuroImage in 2013 (volume 80) (e.g., Van Essen et al. 2013) and similar was done for the Adolescent Brain Cognitive Development (ABCD) consortium in Developmental Cognitive Neuroscience in 2018 (volume 32) (e.g., Casey et al.2018). While it is relatively easy to use these datasets in your own research, I think it is also important to be aware of the considerations that were made when when they were developed. For instance with the ABCD study, it may be useful to further consider how the physical and mental health assessments were chosen (Barch et al. 2018) as well as the ethical considerations that were made, since the study involves the multi-site recruitment of children and adolescents (Clark et al. 2018). For examples of the considerations that may need to be made when collecting data with a clinical sample, see Ye et al. (2019). Its additionally important to evaluate how other data collection considerations, such as the MR sequences used, may influence analyses, e.g., multiband sequences improve temporal resolution, but can also introduce slice-leakage artifacts (Todd et al. 2016; Risk et al. 2018; McNabb et al. 2020). (For a critical discussion, see Longo and Drazen (2016).)
Large Open-Access Neuroimaging Datasets
Over the last two decades, but particularly in recent years, many large open-access neuroimaging datasets have become available (e.g., Marcus et al. 2007; Jack et al. 2008; Van Essen et al. 2013; Hanke et al. 2014; Zuo et al. 2014; Poldrack et al. 2015; Alexander et al. 2017; Taylor et al. 2017; Harms et al. 2018; Casey et al.2018; Milham et al. 2020; Pinho et al. 2020; Nastase et al. 2020). In Table 1, I provide an overview of many of these, spanning multimodal investigations of young adults, lifespan studies of development and/or aging, highly sampled individuals, patient samples, as well as datasets of non-human neuroimaging. Here I have focused on relatively large, novel, or otherwise popular datasets. For instance, OpenNeuro (formerly OpenfMRI) has recently surpassed 500 public datasets, however, most of these are ‘conventional’ in scale (e.g., < 40 participants, single session, sparse additional non-imaging data) and, as such, are not included in the table. These are, of course, still very useful, but their smaller scale makes their applications more limited than the datasets emphasised in this overview. Several schizophrenia datasets are included in the table that are part of SchizConnect (Wang et al. 2016), however, these are also listed separately since they are in federated databases and are otherwise disparate and heterogeneous.
While these datasets are all considered open-access, there is variation in how easy it is to get access to the data. Based on the level of effort required to access the data, I have here coded them each with an “accessibility score” on a 4-point scale: (1) minimal data use agreement required, automatic approval (e.g., IXI, OASIS1, ABIDE, COBRE); (2) some study-specific terms in agreement–to be read carefully, still automatic approval (e.g., HCP, GSP); (3) applications manually approved, often requiring a brief application including a study plan or analysis proposal (e.g., ADNI, CamCAN); (4) more extensive data-use application, requiring institutional support and/or lawyer involvement (e.g., ABCD, HBN). Some datasets have been coded with multiple scores, in cases where some data is shared more readily, but additional variables are provided under restricted terms. As an example, the Human Connectome Project (HCP) is coded as ‘2,3,4.’ While the HCP data is readily shared, it does involve some specific terms, such as not using participant IDs publicly, such as in publications (e.g., including in figures). Additional restricted data (e.g., medical family history) are available under formal application; moreover, genetic data is overseen through NIH dbGaP (‘the Database of Genotypes and Phenotypes’) and requires institutional supporting paperwork and approval.
Consideration is needed when combining data from multiple sites or datasets. It is well-established that there are site effects in MRI in a variety of derived measures. Hagiwara et al. (2020) provide a useful overview of statistics for comparing related measurements, as well as of common imaging-related of variance (e.g., temperature, field nonuniformity, and field strength). Data harmonisation can be attempted at either the initial 3D volume (e.g., signal-to-noise ratio) or specific derived measures (e.g., mean and variance of mean cortical thickness–for instance, using normalised residuals, subsequently combined across sites using site-specific scaling factors) with the goal of matching dataset descriptive statistics. The specific goals of harmonising are important to evaluate. For instance, two sites may exhibit age-related differences in mean cortical thickness, but have different within-site average estimates and age-related slopes. This could be due to site-specific differences in estimated tissue contrast and thus carry forward to the subsequent tissue segmentation and surface reconstruction. Estimates can be adjusted using within-site normalisation along with a linear combination of the site-specific scaling factors. More complex approaches for unseen data are being developed, with between-site harmonisation serving as an active field of methods development, particularly as the availability of open-access datasets continues to increase.
When providing an overview of these large-scale datasets, in addition to crediting the data generators themselves (Pierce et al. 2019), it is also important to acknowledge the software infrastructure that supports them (Ince et al. 2012; Barba et al. 2019). Many of these projects rely on software packages such as the Extensible Neuroimaging Archive Toolkit (XNAT) (Marcus et al. 2007; Herrick et al. 2016)–which was adapted into ConnectomeDB for the HCP (Marcus et al. 2011; Hodge et al. 2016), Collaborative Informatics Neuroimaging Suite (COINS) (Scott et al. 2011; Landis et al. 2016), Longitudinal Online Research and Imaging System (LORIS) (Das et al. 2012), or other online infrastructure such as the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC) (Kennedy et al. 2016), the International Neuroimaging Datasharing Initiative (INDI) (Mennes et al. 2013), the Laboratory of Neuro Imaging (LONI) (Crawford et al. 2016), and OpenNeuro (formerly OpenfMRI) (Poldrack et al. 2013; Poldrack and Gorgolewski 2017). These generally are ‘behind-the-scenes,’ but the data sharing and future analyses from these datasets is as dependent on these software packages as they are on the MRI scanners themselves. While small-scale, within-lab projects can proceed without these packages, they become integral when MRI data is being shared with large groups of users and metadata is linked closely to the individual MRI volumes. Moreover, shared data is often evaluated with some quality control as an initial preprocessing when shared, such as MRIQC (Esteban et al. 2017), fMRIprep (Esteban et al. 2019), PreQual (Cai et al. 2020), or Mindcontrol (Keshavan et al. 2018). Initiatives such as Open Brain Consent (Bannier et al. 2020) are also critical in making neuroimaging data more readily shared (also see Brakewood and Poldrack 2013; Shenkin et al. 2017; White et al. 2020). As a field, we also need to consider long-term data preservation; some previous repositories have become no longer accessible (e.g., fMRI Data Center [fMRIDC] and Biomedical Informatics Research Network [BIRN]) (Horn et al. 2001; Horn and Gazzaniga 2013; Helmer et al. 2011; Hunt 2019).
The overall approach here is ‘scan once, analyse many’ (adapted from the adage ‘write once, read many’ used to describe permanent data storage devices), and as such the benefits of streamlining of the data access process, such as due to these outlined software packages and sharing initiatives, benefits hundreds of ‘secondary analysis’ research groups. For instance, beyond sharing the primary data as a ‘data generator,’ openly sharing quality control (QC), preprocessed data, and data annotations (e.g., manual segmentations) saves others from repeating those efforts.
Example Use Cases
Many innovative studies have already been conducted solely using data from one or more of the datasets outlined in Table 1. Here I provide some examples of this work, to help inspire and demonstrate what can be done using these large-scale open-access neuroimaging datasets. Four general categories of such studies, which particularly benefit from the opportunities created by large datasets, include studies of individual-difference analyses, robust findings, reproducability, and novel methodological findings that may not have been feasible to assess without using existing data. For more exhaustive lists of use cases for these databases, check their respective websites as many of them maintain lists of publications that have relied on their data.
Individual Differences
Many of the findings presented in these example use cases could not have been established in ‘regular’ studies with conventional sample sizes. In particular, studies of individual differences require even larger sample sizes than for within-subject or group differences, and thus particularly benefit from the large-scale of these datasets. Functional connectivity analyses based on network graph-theory methods have become a prominent approach to examine individual differences, and this has been largely reliant on the availability of high-quality fMRI data from large samples (Yeo et al. 2014; Finn et al. 2015; Gratton et al. 2018; Greene et al. 2018; Greene et al. 2020; Seitzman et al. 2019; Cui et al. 2020; Salehi et al. 2020). Spronk et al. (2021) examined functional connectivity in several psychiatric conditions using open datasets (ADHD-200, ABIDE, COBRE), only finding subtle differences in network structure relative to healthy individuals.
The consideration of sex differences in neuroscience is being increasingly discussed, i.e., ‘sex as a biological variable (SABV)’ (Bale and Epperson 2016; Podcasy and Epperson 2016). This is a particularly fitting use of large, open-access neuroimaging datasets, as the inclusion of sex as a factor is unlikely to be an issue given the large sample sizes. Forde et al. (2020) examined sex differences in brain structure across the lifespan, using PNC, HCP, and OASIS-3 datasets (N = 3069). They observed an interaction, where males had less within-region variability in early years, but more variability in later years. This result extended previous work that had looked at narrower age ranges, such as Wierenga et al. (2018) with PING, more coarse brain size measures (e.g., van der Linden et al. 2017, with HCP), or more specific regions (e.g., van Eijk et al. 2020, with the hippocampus). Other studies have used these datasets to examine sex-related differences in brain activity or functional connectivity (e.g., Scheinost et al. 2015; Dumais et al. 2018; Dhamala et al. 2020; Li et al. 2020). A handful of studies have also examined brain structure or function differences in relation to personality traits (Riccelli et al. 2017; Gray et al. 2018; Nostro et al. 2018; Owens et al. 2019; Sripada et al. 1900). Some results indicate that personality should be examined separately for each sex (Nostro et al. 2018); many results appear replicable, but effects are relatively weak.
Examining age-related differences in brain structure and function has become a prominent topic in studies that use large open-access datasets. Some of this work is described below, in the methods development section, as it was associated with the development of novel methods. Additionally, using the movie watching data from CamCAN, Geerligs and Campbell (2018) examined differences in inter-participant synchrony and found age-related differences in how shared functional networks were activated, corresponding to processing of naturalistic experiences. In a subsequent study, Reagh et al. (2020) examined the same movie-watching fMRI data and observed increases in posterior (but not anterior) hippocampal activity in relation to event boundaries, but also that these increases were attenuated by aging (also see Ben-Yakov and Henson 2018).
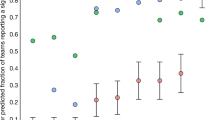
Several studies have been investigating individual differences in global fMRI signal (and the potential utility and limitations of global signal regression). This includes examining differences in relation to scan acquisitions and psychiatric conditions (Power et al. 2017), as well as behavioural features (Li et al. 2019), as shown in Fig. 2b (also see Smith et al. 2015). Others have examined the reproducability of individual differences and how data collection is influenced by multi-site factors, such as in the ABIDE (Abraham et al. 2017) and ABCD (Marek et al. 2019) studies. Within highly-sampled individuals, detailed network analyses can be conducted for each participant (Gordon et al. 2017) (Fig. 3a) and intra-individual differences can also be examined, such as the influence of caffeine on functional connectivity (Poldrack et al. 2015) (Fig. 3b) and BOLD signal variability (Yang et al. 2018).
Inter- and intra-individual differences in functional connectivity from highly-sampled individuals. a Inter-individual variability across 10 individuals, reprinted from Gordon et al. (2017). b Intra-individual variability (related to fasting/caffeination), reprinted from Poldrack et al. (2015). Distinct colours denote each functional network. Arrows highlight specific regions of inter-individual variability
In addition to using individual difference measures as continuous measures, some studies have used large-scale datasets to characterise potential subtypes within patient samples. Using different analytical approaches, Dong et al. (2017) and Zhang et al. (2016) demonstrated heterogeneity and subtypes in atrophy patterns due to Alzheimer’s disease using data from ADNI. Guo et al. (2020) also examined subtypes in the ADNI dataset, but instead focused on those with mild cognitive impairment. Furthermore, large open-access datasets have also been used to develop subtyping methods for autism (Easson et al. 2019; Tang et al. 2020) and schizophrenia (Castro-de-Araujo et al. 2020).
Other studies have examined differences in structure. For instance, Holmes et al. (2016) used the GSP dataset and examined relationships between cortical structure and different individual difference measures of cognitive control (e.g., sensation seeking, impulsivity, and substance use [alcohol, caffeine, and cigarettes]). Cao et al. (2017) examined gyrification trajectories across the lifespan, from 4 to 83, and in relation to several psychiatric conditions (major depression disorder, bipolar disorder, and schizophrenia), by combining data from a within-lab sample with the NKI and COBRE datasets.
A narrow, but particularly beneficial use of these large datasets is to examine the frequency of infrequent brain morphological features. In conducting a conventional study, Weiss et al. (2020) identified two participants with no apparent olfactory bulbs, despite no impairments in olfactory performance. To examine the prevalence in the general population, the researchers examined the structural MRIs of 1113 participants from the HCP study. Three participants were identified–all had monozygotic twins that had visible olfactory bulbs. MRIs from one set of twins is shown in Fig. 4a. Moreover, the twins without apparent olfaction bulbs had higher olfaction scores than their twins with visible bulbs. A fourth participant was also identified, though the MRI was sufficiently blurry that it is difficult to be confident if the olfactory bulb is present. All identified individuals without apparent olfactory bulbs were women, occurring in 0.6% of women, with an increased likelihood in left-handed women (4.3%). Another morphological variation examined in large datasets is the incomplete hippocampal inversion, sometimes referred to a hippocampal malrotation, as shown in Fig. 4b. This can be identified by the diameter and curvature of the hippocampus, as well as angle in relation to the parahippocampal gyrus (see Caciagli et al.2019). In a large study of 2008 participants (Cury et al. 2015), the presence of this anatomical feature was visible in 17% of left hippocampus and 6% of right hippocampus. Cury et al. (2020) replicated this result in PING, finding a similar incidence rate, as well as examined the genetic predictors. Incomplete hippocampal inversion has been associated with an increased risk of developing epilepsy (Gamss et al. 2009; Caciagli et al. 2019). Other features may be useful to examine in large datasets, but have not been yet, such as the presence of single or double cingulate sulci (Vogt et al. 1995; Cachia et al. 2016; Amiez et al. 2019) and orbitofrontal sulci patterns (Chiavaras and Petrides 2000; Nakamura et al. 2007; Li et al. 2019). Heschl’s gyrus morphology variants have been examined in one large dataset (Marie et al. 2016), but would benefit from further research.
Examples of infrequent morphological features examined in large datasets. a Typical olfactory bulbs and no apparent bulbs in monozygotic twins, shown on a T2-weighted coronal image, adapted from Weiss et al. (2020). b Typical hippocampus and incomplete hippocampal inversion, shown on a T1-weighted coronal image, adapted from Caciagli et al. (2019). c Single and double cingulate sulcus, shown on the medial view of a reconstructed cortical surface, adapted from Cachia et al. (2016).
Robust Findings
Significant results based on large sample sizes are less likely to be due to random chance, and thus can be considered more robust (though admittedly can still occur due to systematic error) (Hung et al. 1997; Thiese et al. 2016; Madan 2016; Greenland 2019). Several studies have used the HCP task-fMRI data to evaluate the robustness of task condition contrasts and functional connectivity configurations (Barch et al. 2013; Shine et al. 2016; Schultz and Cole 2016; Shah et al. 2016; Westfall et al. 2017; Zuo et al. 2017; Nickerson 2018; Markett et al. 2020; Jiang et al. 2020). Margulies et al. (2016) used the HCP data to demonstrate gradients within default-mode network structure, providing significant insights into how sensory and association cortices communicate (Fig. 2a). Providing an example from a patient dataset, Cousineau et al. (2017) observed differences in white matter fascicles associated with Parkinson’s disease using the PPMI dataset, a result that was strengthened by the relatively large sample size of the dataset and the acquisition of test-retest DTI scans. See Fig. 5 for a summary of white-matter tracts.
Overview of white-matter tracts. Reprinted from Thiebaut de Schotten et al. (2015)
As an increasing number of datasets collect movie watching data, this additionally affords the opportunity of bringing hyperalignment methods to the mainstream. Of course, movie watching/naturalistic stimuli themselves are able to provide insights into brain function in new ways that resting-state and task fMRI methods could not. That said, Haxby and colleagues have demonstrated that hyperalignment should be considered as an alternative to conventional anatomical-based normalisation. Briefly, conventional fMRI methods rely on than warping the structural MRI into a common space and then applying that non-linear transform to co-registered rest/task fMRI data. In contrast, hyperalignment uses the time-varying activations related to a movie-watching stimuli as a common, high-dimensional representation to serve as the transformation matrix to bring individuals into a common MRI space (Fig. 6c). A comparison of alignment methods is shown in Fig. 6a (task data from an six-category animal localiser shown). Despite comparable performance within-subject, conventional anatomical methods perform poorly across subjects. Analyses indicated that 10:25 min (250 TRs) of movie watching was sufficient for hyperalignment methods (Haxby et al. 2011; Guntupalli et al. 2016). More recent development of the connectivity hyperalignment method–as opposed to the original approach, now termed ‘response hyperalignment’–have improved the utility of the method in aligning connectivity data, but response hyperalignment nonetheless is advantageous in some situations (Guntupalli et al. 2018; Haxby et al. 2020).
Performance of hyperalignment in comparison to conventional anatomical alignment. a Classification performance from a six-category animal localiser. wsMVPC denotes within-subject multivariate pattern classification; bsMVPC denotes between-subject. b Between-subject MVPC performance of movie-watching data, as a function of amount of data used in the hyperalignment. c Illustration of the method. Reprinted from Guntupalli et al. (2016)
Reproducability
An added benefit of analysing public data is that outputs can be compared directly. With some datasets, this is the primary function of the dataset, such as with the 7 T test-retest (Gorgolewski et al. 2013) and NARPS datasets (Botvinik-Nezer et al. 2019; Botvinik-Nezer et al. 2020). With these datasets, researchers can test their ability to reproduce the analysis pipeline or develop new analysis pipelines and compare the output with previous results as benchmarks (i.e., analytical flexibility) (also see Silberzahn et al. 2018; Schweinsberg et al. 2021). This allows for confidence that the same input data was used, rather than an attempt to collect new data and replicate prior results. Some studies have taken this one step further, implementing a ‘multiverse’ approach in examining data through multiple methods, by the same research group (Carp (2012) and Pauli et al. (2016); also see Steegen et al. (2016) and Botvinik-Nezer et al. (2020)). In a similar vein, a recent large-scale collaboration used a subset of the HCP data to assess consistency across tractography segmentation protocols (Schilling et al. 2020). Here it was a clear benefit that public data that all researchers could access was already available (also see ADNI TADPOLE challenge: Marinescu et al. 2020).
A related use is more teaching-oriented. Since the MRI data from these datasets are publicly available–at least after agreeing to the initial data-use terms. As such, they can readily be used as specific real-world examples of acquired data. To provide a concrete example of this, I made Fig. 7 to show instances of MRI artifacts using data from ABIDE. While this figure itself should be useful for those familiarising themselves with the neuroimaging data, I have included the participant IDs to allow interested readers to go one step further and examine the same MRI volumes that I used to make the figures.
Examples of MRI artifacts in T1 volumes present in the ABIDE dataset. a Head motion artifacts, with increasing magnitude of motion left to right. Volumes comparable to images 1 and 2 would be suitable for further analysis, but those rated as 3 through 5 have too much head motion to be useable. While most of the ABIDE data is of reasonable quality, it is large dataset and includes participants with autism spectrum disorder as well as children, both factors known to be associated with increased head motion (Pardoe et al. 2016; Engelhardt et al. 2017; Greene et al. 2018). b Ghosting artifacts, visible as overlapping images. The example on the left is only visible in the background with a constrained intensity range, but still results in distortions in the image. The image on the right shows a clear duplicate contour of the back of the head. c Blood flow artifact, creating a horizontal band of distortion, here affecting temporal lobe imaging. d Spike noise artifact, resulting in inconsistent signal intensity. e Coil failure artifact, resulting in a regional distortion around the affected coil. Participant IDs are included below each image to allow for the further examination of the original 3D volumes. Artifact MRIs were identified with the aid of MRIQC (Esteban et al. 2017). Pre-computed results are available from https://mriqc.s3.amazonaws.com/abide/T1w_group.html
A further advantage of the large sample sizes available in many of the featured datasets is that they allow for cross-validation analyses, where analyses are conducted on subsets of the data and patterns of results can be evaluated as being replicable, particularly across multiple sites. Among other rigorous analyses, Abraham et al. (2017) use data from ABIDE and examine inter-site cross validation, where data is pooled across several sites and used to predict autism spectrum disorder diagnosis in other sites (also see Varoquaux 2018; Owens et al. 2019). Several age-prediction studies have similarly used several datasets to identify age-sensitive regions and predict age in independent datasets (e.g., Cole et al. 2015; Madan and Kensinger 2018; Bellantuono et al. 2021). This has been done with other topics as well, where large datasets such as HCP and ADNI are used as replication samples and works particularly well for studies that are otherwise examining individual differences (e.g., Hodgson et al. 2017; Madan & Kensinger, 2017a; Madan, 2019b; Richard et al. 2018; Young et al. 2018; Grady et al. 2020; Baranger et al. 2020; Baranger et al. 2020; Kharabian Masouleh et al. 2020; Weiss et al. 2020; Yang et al. 2020; van Eijk et al. 2020). See Fig. 8 for an overview of anatomical-based cortical parcellations.
Overview of cortical parcellation approaches instantiated in FreeSurfer. Parcellations are shown on inflated and pial surfaces and an oblique coronal slice, reconstructed from an MRI of a young adult. Updated from Madan and Kensinger (2018) to include Collantoni et al. (2020) and more clearly show parcellation boundaries on the inflated surface; visualisations produced based on previously described methods (Madan and Kensinger 2016; Klein and Tourville 2012; Destrieux et al. 2010; Scholtens et al. 2018; Fan et al. 2016; Hagmann et al. 2008)
Some studies have examined how results vary in relation to sample size, either in the form of a meta-analysis or through the analysis of subsets of the data, see Fig. 9 (Termenon et al. 2016; Varoquaux 2018; Zuo et al. 2019; Grady et al. 2020; also see Schönbrodt and Perugini 2013). It is well-established, unfortunately, that smaller cohorts often result in overestimation of effect sizes (Hullett and Levine 2003; Forstmeier and Schielzeth 2011; Varoquaux 2018). Larger datasets should result in more accurate effect sizes and, in principle, should yield more robust and generalisable findings. By necessity, larger datasets include more heterogeneous data than smaller datasets. Admittedly, the use of large datasets makes most analyses yield either clearly significant or non-significant results, due to the large sample sizes, thus making the distinction between the practical relevance or meaningful effect size important to consider (e.g., ‘smallest effect size of interest’; Lakens et al. 2018), rather than the statistical significance itself.
Reported prediction accuracy as a function of sample size for studies in different meta-analyses. Reprinted from Varoquaux (2018). Copyright 2018, Elsevier
Methods Development
Some uses of large open-access datasets have been for purposes that would not have been practical as the primary outcome of new data collection, but take advantage of existing data to refine analysis methods going forward. For example, Esteban et al. (2017) developed MRIQC to automatically and quantitatively assess MR image quality on a variety of metrics, using data from ABIDE and LA5c. Advanced Normalisation Tools (ANTs) (Tustison et al. 2014) is a volumetric pipeline for image registration, tissue segmentation, and cortical thickness estimation, among other structural MRI operations. This 2014 paper that demonstrating a rigourous evaluation of this comprehensive pipeline uses four open-access datasets (IXI, MMRR, NKI, OASIS1) to showcase the robustness of the pipeline, including example figures corresponding to specific individual MRI inputs. Davis (2021) recently used data from OASIS1 to examine variability in cortical depth (i.e., distance from the scalp to the cortical surface, through the skull) as a means of assessing regional variability for transcranial stimulation research. IXI and OASIS1 have been used as a training dataset for a large number of methodological developments, especially in relation to age-related effects (e.g., Schrouff et al. 2013; Yun et al. 2013; Romero et al. 2015; Auzias et al. 2015; Wang et al.2016). In another instance, Madan (2019a) developed a novel toolbox for quantifying sulcal morphology and evaluated the generalisability of the method across several healthy aging dataset–OASIS1 and DLBS–as well as SALD as a non-Western sample and CCBD to assess test-retest reliability. HCP, GSP, MASSIVE, and Maclaren et al. (2014) have also useful for assessing test-retest reliability (also see Madan and Kensinger2017b).
Madan (2018) used data from CamCAN to replicate a number of findings that have been previously shown, including increased head motion in older adults, decreases in head motion associated with movie watching, and weak but statistically significant effects of head motion on estimates of cortical morphology (Fig. 10). This lead to the proposal that watching a movie during the acquisition of a structural volume would improve data quality, though consideration is needed, e.g., this would be problematic if the structural volume was then followed by a resting-state sequence. Body–mass index (BMI) was also associated with increased respiratory-related apparent head motion, determined through the use of multiple estimates of head motion, and has since been supported by a further study that also used these large-scale datasets, Power et al. (2019). While the focus of Madan (2018) was aging effects on head motion, Power et al. (2019) examined head motion effects on fMRI signal, using the HCP, GSP, and MyConnectome datasets (complemented by additional within-lab datasets). Pardoe et al. (2016) have examined head motion effects across several clinical populations using data from ABIDE, ADHD-200, and COBRE; Zacà et al. (2018) examined head motion in the PPMI.
Correlations between head-motion during rest and movie-watching fMRI scans with age and body-mass index (BMI). Head motion axes are log-10 scaled to better show inter-individual variability. Reprinted from Madan (2018)
Using data from ADNI, King et al. (2009, 2010) demonstrated that fractal dimensionality can be a more sensitive measure of brain structure differences associated with Alzheimer’s disease than conventional measures of cortical thickness and gyrification. Inspired by this work, Madan and Kensinger (2016) examined age-related differences in the IXI database and found this as well; later using IXI, OASIS1, and a within-lab sample to examine subcortical structure (Madan and Kensinger 2017a) (Fig. 11). Several later studies expanded on these initial findings (Madan and Kensinger 2017b, 2018; Madan 2018, 2019b, 2021), fully reliant on large open-access datasets.
Closing Thoughts
Conducting a new neuroimaging study can easily have a budget upwards of 20,000 dollars (or pounds) for the MRI scan time, let alone the time and labour associated with participants and researchers involved. The value of already collected data grows greatly in the unlikely circumstances of a wide spread public health issue (as in our current COVID-19 pandemic), where contact between individuals must be minimised, but PhD training–as well as furthering our understanding of the brain–must continue.
In using these large-access open-access datasets, we must consider the decisions that went into the data we are now using. Some of these are still yet to be made, such as how to harmonise the MRI data from multiple sites or similarly how to reconcile potential differences in screening criteria between sites (particularly when data is aggregated at a later stage, rather than a planned multi-site study). Other decisions have already been made and simply need to be incorporated into the subsequent research despite limitations, such as the artifacts in the multiband sequence used in the HCP (Risk et al. 2018; McNabb et al. 2020) and specifics of the task design used in the ABCD study (Bissett et al. 2020). We also need to consider the prior use of the datasets, specifically to become over-reliant, and thus over-fit, our knowledge as a field to specific datasets. Given the current state of the field, this is of particular concern with HCP and ADNI–if too many analyses are based on these specific samples, that may bias our understanding of the brain. As an example, it is worth re-visiting the recruitment procedures for these studies and evaluating how representative they are of the desired population we may want to generalise or what sampling biases may be present, e.g., education level, socioeconomic status, response bias. For further, more focused discussions on current topics and using open-access neuroimaging datasets in specific contexts, please see the referenced papers: development (Gilmore 2016; Klapwijk et al. 2021), aging (Reagh and Yassa 2017), brain morphology (Madan 2017), naturalistic stimuli (Vanderwal et al. 2019; Finn et al. 2020; DuPre et al. 2020), head motion (Ai et al. 2021), non-human primates (Milham et al. 2020) data management (Borghi and Van Gulick, A. E. 2018), computational reproducibility (Kennedy et al. 2019; Poldrack et al. 2017; Poldrack et al. 2019; Carmon et al. 2020), and machine learning (Dwyer et al. 2018).
As a final set of remarks, I would like to direct readers to several articles to help deepen how they make think of the brain. Though we hopefully have sufficiently moved on from the days of circular inferences, Vul et al. (2009) remains an important article for those entering the field. Weston et al. (2019) raises many important considerations associated with working with secondary datasets, while Broman and Woo (2017) and Wilson et al. (2017) are essential reads for data organisation and scientific computing, respectively–both critical topics when working with large datasets. Pernet and Madan (2020) provides guidance for producing visualisations of MRI analyses. Eickhoff et al. (2018) and Uddin et al. (2019) provide insightful discussions for thinking about the structure of the brain.
References
Abraham, A., Milham, M.P., Martino, A.D., Craddock, R.C., Samaras, D., Thirion, B., & Varoquaux, G. (2017). Deriving reproducible biomarkers from multi-site resting-state data: An autism-based example. NeuroImage, 147, 736–745. https://doi.org/10.1016/j.neuroimage.2016.10.045.
ADHD-200 Consortium. (2012). The ADHD-200 consortium: a model to advance the translational potential of neuroimaging in clinical neuroscience. Frontiers in Systems Neuroscience, 6, 62. https://doi.org/10.3389/fnsys.2012.00062.
Ai, L., Craddock, R.C., Tottenham, N., Dyke, J.P., Lim, R., Colcombe, S., & Franco, A.R. (2021). Is it time to switch your t1w sequence? assessing the impact of prospective motion correction on the reliability and quality of structural imaging. NeuroImage, 226, 117585. https://doi.org/10.1016/j.neuroimage.2020.117585.
Aine, C.J., Bockholt, H.J., Bustillo, J.R., Cañive, J. M., Caprihan, A., Gasparovic, C., & Calhoun, V.D. (2017). Multimodal neuroimaging in schizophrenia: Description and dissemination. Neuroinformatics, 15, 343–364. https://doi.org/10.1007/s12021-017-9338-9.
Alexander, L.M., Escalera, J., Ai, L., Andreotti, C., Febre, K., Mangone, A., & Milham, M.P. (2017). An open resource for transdiagnostic research in pediatric mental health and learning disorders. Scientific Data, 4, 170181. https://doi.org/10.1038/sdata.2017.181.
Aliko, S., Huang, J., Gheorghiu, F., Meliss, S., & Skipper, J.I. (2020). A naturalistic neuroimaging database for understanding the brain using ecological stimuli. Scientific Data. 7, 347. https://doi.org/10.1038/s41597-020-00680-2.
Allen, E.J., St-Yves, G., Wu, Y., Breedlove, J.L., Dowdle, L.T., Caron, B., & Kay, K. (2021). A massive 7 T fMRI dataset to bridge cognitive and computational neuroscience. bioRxiv. https://doi.org/10.1101/2021.02.22.432340.
Amiez, C., Sallet, J., Hopkins, W.D., Meguerditchian, A., Hadj-Bouziane, F., Hamed, S.B., & Petrides, M. (2019). Sulcal organization in the medial frontal cortex provides insights into primate brain evolution. Nature Communications, 10, 3437. https://doi.org/10.1038/s41467-019-11347-x.
Amunts, K., Lepage, C., Borgeat, L., Mohlberg, H., Dickscheid, T., Rousseau, M.-E., & Evans, A.C. (2013). Bigbrain: An ultrahigh-resolution 3D human brain model. Science, 340(6139), 1472–1475. https://doi.org/10.1126/science.1235381.
Armstrong, N.M., An, Y., Beason-Held, L., Doshi, J., Erus, G., Ferrucci, L., & Resnick, S.M. (2019). Sex differences in brain aging and predictors of neurodegeneration in cognitively healthy older adults. Neurobiology of Aging, 81, 146–156. https://doi.org/10.1016/j.neurobiolaging.2019.05.020.
Auzias, G., Brun, L., Deruelle, C., & Coulon, O. (2015). sulcal landmarks: Algorithmic and conceptual improvements in the definition and extraction of sulcal pits. NeuroImage, 111, 12–25. https://doi.org/10.1016/j.neuroimage.2015.02.008.
Babayan, A., Erbey, M., Kumral, D., Reinelt, J.D., Reiter, A.M.F., Röbbig, J., & Villringer, A. (2019). A mind-brain-body dataset of MRI, EEG, cognition, emotion, and peripheral physiology in young and old adults. Scientific Data, 6, 180308. https://doi.org/10.1038/sdata.2018.308.
Bale, T.L., & Epperson, C.N. (2016). Sex as a biological variable: Who, what, when, why, and how. Neuropsychopharmacology, 42, 386–396. https://doi.org/10.1038/npp.2016.215.
Bannier, E., Barker, G., Borghesani, V., Broeckx, N., Clement, P., De la Iglesia Vaya, M, & Zhu, H. (2020). The Open Brain Consent: Informing research participants and obtaining consent to share brain imaging data. PsyArXiv. https://doi.org/10.31234/osf.io/f6mnp.
Baranger, D.A.A., Demers, C.H., Elsayed, N.M., Knodt, A.R., Radtke, S.R., Desmarais, A., & Bogdan, R. (2020). Convergent evidence for predispositional effects of brain gray matter volume on alcohol consumption. Biological Psychiatry, 87, 645–655. https://doi.org/10.1016/j.biopsych.2019.08.029.
Baranger, D.A.A., Few, L.R., Sheinbein, D.H., Agrawal, A., Oltmanns, T.F., Knodt, A.R., & Bogdan, R. (2020). Borderline personality traits are not correlated with brain structure in two large samples. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 5, 669–677. https://doi.org/10.1016/j.bpsc.2020.02.006.
Barba, L.A., Bazán, J., Brown, J., Guimera, R.V., Gymrek, M., Hanna, A., & Yehudi, Y. (2019). Giving software its due through community-driven review and publication. OSF Preprints. https://doi.org/10.31219/osf.io/f4vx6.
Barch, D.M., Albaugh, M.D., Avenevoli, S., Chang, L., Clark, D.B., Glantz, M.D., & Sher, K.J. (2018). Demographic, physical and mental health assessments in the adolescent brain and cognitive development study: Rationale and description. Developmental Cognitive Neuroscience, 32, 55–66. https://doi.org/10.1016/j.dcn.2017.10.010.
Barch, D.M., Burgess, G.C., Harms, M.P., Petersen, S.E., Schlaggar, B.L., Corbetta, M., & Van Essen, D.C. (2013). Function in the human connectome: Task-fMRI and individual differences in behavior. NeuroImage, 80, 169–189. https://doi.org/10.1016/j.neuroimage.2013.05.033.
Bellantuono, L., Marzano, L., Rocca, M.L., Duncan, D., Lombardi, A., Maggipinto, T., & Bellotti, R. (2021). Predicting brain age with complex networks: From adolescence to adulthood. NeuroImage, 225, 117458. https://doi.org/10.1016/j.neuroimage.2020.117458.
Bellec, P., & Boyle, J.A. (2019). Bridging the gap between perception and action: the case for neuroimaging, AI and video games. PsyArXiv. https://doi.org/10.31234/osf.io/3epws.
Bellec, P., Chu, C., Chouinard-Decorte, F., Benhajali, Y., Margulies, D.S., & Craddock, R.C. (2017). The Neuro Bureau ADHD-200 preprocessed repository. NeuroImage, 144, 275–286. https://doi.org/10.1016/j.neuroimage.2016.06.034.
Bennett, D.A., Schneider, J.A., Buchman, A.S., Barnes, L.L., Boyle, P.A., & Wilson, R.S. (2012). Overview and findings from the Rush memory and aging project. Current Alzheimer Research, 9, 646–663. https://doi.org/10.2174/156720512801322663.
Ben-Yakov, A., & Henson, R.N. (2018). The hippocampal film editor: Sensitivity and specificity to event boundaries in continuous experience. Journal of Neuroscience, 38, 10057–10068. https://doi.org/10.1523/jneurosci.0524-18.2018.
Bertram, L., Böckenhoff, A., Demuth, I., Düzel, S., Eckardt, R., Li, S.-C., & Steinhagen-Thiessen, E. (2014). Cohort profile: The Berlin Aging Study II (BASE-II). International Journal of Epidemiology, 43, 703–712. https://doi.org/10.1093/ije/dyt018.
Bissett, P.G., Hagen, M.P., & Poldrack, R.A. (2020). A cautionary note on stop-signal data from the adolescent brain cognitive development [ABCD] study. bioRxiv. https://doi.org/10.1101/2020.05.08.084707.
Bookheimer, S.Y., Salat, D.H., Terpstra, M., Ances, B.M., Barch, D.M., Buckner, R.L., & Yacoub, E. (2019). The Lifespan Human Connectome Project in Aging: An overview. NeuroImage, 185, 335–348. https://doi.org/10.1016/j.neuroimage.2018.10.009.
Borghi, J.A., & Van Gulick, A.E. (2018). Data management and sharing in neuroimaging: Practices and perceptions of MRI researchers. PLOS ONE, 13, e0200562. https://doi.org/10.1371/journal.pone.0200562.
Botvinik-Nezer, R., Holzmeister, F., Camerer, C.F., Dreber, A., Huber, J., Johannesson, M., & Schonberg, T. (2020). Variability in the analysis of a single neuroimaging dataset by many teams. Nature, 582, 84–88. https://doi.org/10.1038/s41586-020-2314-9.
Botvinik-Nezer, R., Iwanir, R., Holzmeister, F., Huber, J., Johannesson, M., Kirchler, M., & Schonberg, T. (2019). fMRI data of mixed gambles from the neuroimaging analysis replication and prediction study. Scientific Data, 6, 106. https://doi.org/10.1038/s41597-019-0113-7.
Brain Development Cooperative Group, & Evans, A.C. (2006). The NIH MRI study of normal brain development. NeuroImage, 30, 184–202. https://doi.org/10.1016/j.neuroimage.2005.09.068.
Brakewood, B., & Poldrack, R.A. (2013). The ethics of secondary data analysis: Considering the application of Belmont principles to the sharing of neuroimaging data. NeuroImage, 82, 671–676. https://doi.org/10.1016/j.neuroimage.2013.02.040.
Breitner, J.C.S., Poirier, J., Etienne, P.E., & The PREVENT-AD Research Group, J.M.L. (2016). Rationale and structure for a new center for Studies on Prevention of Alzheimer’s Disease (stop-ad). Journal of Prevention of Alzheimer’s Disease, 3, 236–242. https://doi.org/10.14283/JPAD.2016.121.
Broman, K.W., & Woo, K.H. (2017). Data organization in spreadsheets. The American Statistician, 72, 2–10. https://doi.org/10.1080/00031305.2017.1375989.
Button, K.S., Ioannidis, J.P.A., Mokrysz, C., Nosek, B.A., Flint, J., Robinson, E.S.J., & Munafò, M. R. (2013). Power failure: why small sample size undermines the reliability of neuroscience. Nature Reviews Neuroscience, 14, 365–376. https://doi.org/10.1038/nrn3475.
Cachia, A., Borst, G., Tissier, C., Fisher, C., Plaze, M., Gay, O., & Raznahan, A. (2016). Longitudinal stability of the folding pattern of the anterior cingulate cortex during development. Developmental Cognitive Neuroscience, 19, 122–127. https://doi.org/10.1016/j.dcn.2016.02.011.
Caciagli, L., Wandschneider, B., Xiao, F., Vollmar, C., Centeno, M., Vos, S.B., & Koepp, M.J. (2019). Abnormal hippocampal structure and function in juvenile myoclonic epilepsy and unaffected siblings. Brain: A Journal of Neurology, 142, 2670–2687. https://doi.org/10.1093/brain/awz215.
Cai, L.Y., Yang, Q., Hansen, C.B., Nath, V., Ramadass, K., Johnson, G.W., & Landman, B.A. (2020). PreQual: An automated pipeline for integrated preprocessing and quality assurance of diffusion weighted MRI images. bioRxiv. https://doi.org/10.1101/2020.09.14.260240.
Cao, B., Mwangi, B., Passos, I.C., Wu, M.-J., Keser, Z., Zunta-Soares, G.B., & Soares, J.C. (2017). Lifespan gyrification trajectories of human brain in healthy individuals and patients with major psychiatric disorders. Scientific Reports, 7, 511. https://doi.org/10.1038/s41598-017-00582-1.
Cárdenas-de-la-Parra, A., Lewis, J.D., Fonov, V.S., Botteron, K.N., McKinstry, R.C., Gerig, G., & Collins, D.L. (2021). A voxel-wise assessment of growth differences in infants developing autism spectrum disorder. NeuroImage: Clinical, 29, 102551. https://doi.org/10.1016/j.nicl.2020.102551.
Carmon, J., Heege, J., Necus, J.H., Owen, T.W., Pipa, G., Kaiser, M., & Wang, Y. (2020). Reliability and comparability of human brain structural covariance networks. NeuroImage, 220, 117104. https://doi.org/10.1016/j.neuroimage.2020.117104.
Carp, J. (2012). On the plurality of (methodological) worlds: Estimating the analytic flexibility of fMRI experiments. Frontiers in Neuroscience, 6, 149. https://doi.org/10.3389/fnins.2012.00149.
Casey, B., Cannonier, T., Conley, M.I., Cohen, A.O., Barch, D.M., Heitzeg, M.M., & Dale, A.M. (2018). The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Developmental Cognitive Neuroscience, 32, 43–54. https://doi.org/10.1016/j.dcn.2018.03.001.
Castro-de-Araujo, L.F., Machado, D.B., Barreto, M.L., & Kanaan, R.A. (2020). Subtyping schizophrenia based on symptomatology and cognition using a data driven approach. Psychiatry Research: Neuroimaging, 304, 111136. https://doi.org/10.1016/j.pscychresns.2020.111136.
Çetin, M. S., Christensen, F., Abbott, C.C., Stephen, J.M., Mayer, A.R., Cañive, J. M., & Calhoun, V. D. (2014). Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia. NeuroImage, 97, 117–126. https://doi.org/10.1016/j.neuroimage.2014.04.009.
Chang, N., Pyles, J.A., Marcus, A., Gupta, A., Tarr, M.J., & Aminoff, E.M. (2019). BOLD5000, a public fMRI dataset while viewing 5000 visual images. Scientific Data, 6, 49. https://doi.org/10.1038/s41597-019-0052-3.
Chen, B., Xu, T., Zhou, C., Wang, L., Yang, N., Wang, Z., & Weng, X.-C. (2015). Individual variability and test-retest reliability revealed by ten repeated resting-state brain scans over one month. PLOS ONE, 10, e0144963. https://doi.org/10.1371/journal.pone.0144963.
Chen, J., Leong, Y.C., Honey, C.J., Yong, C.H., Norman, K.A., & Hasson, U. (2017). Shared memories reveal shared structure in neural activity across individuals. Nature Neuroscience, 20, 115–125. https://doi.org/10.1038/nn.4450.
Chertkow, H., Borrie, M., Whitehead, V., Black, S.E., Feldman, H.H., Gauthier, S., & Rylett, R.J. (2019). The Comprehensive Assessment of Neurodegeneration and Dementia: Canadian cohort study. Canadian Journal of Neurological Sciences, 46, 499–511. https://doi.org/10.1017/cjn.2019.27.
Chiavaras, M.M., & Petrides, M. (2000). Orbitofrontal sulci of the human and macaque monkey brain. Journal of Comparative Neurology, 422, 35–54. https://doi.org/10.1002/(sici)1096-9861(20000619)422:1<35::aid-cne3>3.0.co;2-e.
Choe, A.S., Jones, C.K., Joel, S.E., Muschelli, J., Belegu, V., Caffo, B.S., & Pekar, J.J. (2015). Reproducibility and temporal structure in weekly resting-state fMRI over a period of 3.5 years. PLOS ONE, 10, e0140134. https://doi.org/10.1371/journal.pone.0140134.
Clark, D.B., Fisher, C.B., Bookheimer, S., Brown, S.A., Evans, J.H., Hopfer, C., & Yurgelun-Todd, D. (2018). Biomedical ethics and clinical oversight in multisite observational neuroimaging studies with children and adolescents: The ABCD experience. Developmental Cognitive Neuroscience, 32, 143–154. https://doi.org/10.1016/j.dcn.2017.06.005.
Cole, J.H., Leech, R., & And, D.J.S. (2015). Prediction of brain age suggests accelerated atrophy after traumatic brain injury. Annals of Neurology, 77, 571–581. https://doi.org/10.1002/ana.24367.
Collantoni, E., Madan, C.R., Meneguzzo, P., Chiappini, I., Tenconi, E., Manara, R., & Favaro, A. (2020). Cortical complexity in anorexia nervosa: A fractal dimension analysis. Journal of Clinical Medicine, 9, 833. https://doi.org/10.3390/jcm9030833.
Cousineau, M., Jodoin, P.-M., Garyfallidis, E., Côté, M.-A., Morency, F. C., Rozanski, V., & Descoteaux, M. (2017). A test-retest study on Parkinson’s PPMI dataset yields statistically significant white matter fascicles. NeuroImage: Clinical, 16, 222–233. https://doi.org/10.1016/j.nicl.2017.07.020.
Crawford, K.L., Neu, S.C., & Toga, A.W. (2016). The image and data archive at the laboratory of neuro imaging. NeuroImage, 124, 1080–1083. https://doi.org/10.1016/j.neuroimage.2015.04.067.
Cui, Z., Li, H., Xia, C.H., Larsen, B., Adebimpe, A., Baum, G.L., & Satterthwaite, T.D. (2020). Individual variation in functional topography of association networks in youth. Neuron, 106, 340–353, e8. https://doi.org/10.1016/j.neuron.2020.01.029.
Cury, C., Scelsi, M.A., Toro, R., Frouin, V., Artiges, E., & Grigis, A. (2020). Genome wide association study of incomplete hippocampal inversion in adolescents. PLOS ONE, 15, e0227355. https://doi.org/10.1371/journal.pone.0227355.
Cury, C., Toro, R., Cohen, F., Fischer, C., Mhaya, A., Samper-González, J., & Colliot, O. (2015). Incomplete hippocampal inversion: a comprehensive MRI study of over 2000 subjects. Frontiers in Neuroanatomy, 9, 160. https://doi.org/10.3389/fnana.2015.00160.
Dagley, A., LaPoint, M., Huijbers, W., Hedden, T., McLaren, D.G., Chatwal, J.P., & Schultz, A.P. (2017). Harvard Aging Brain Study: Dataset and accessibility. NeuroImage, 144, 255–258. https://doi.org/10.1016/j.neuroimage.2015.03.069.
Das, S., Zijdenbos, A.P., Harlap, J., Vins, D., & Evans, A.C. (2012). LORIS: a web-based data management system for multi-center studies. Frontiers in Neuroinformatics, 5, 37. https://doi.org/10.3389/fninf.2011.00037.
Davis, N.J. (2021). Variance in cortical depth across the brain surface: Implications for transcranial stimulation of the brain. European Journal of Neuroscience. https://doi.org/10.1111/ejn.14957.
Destrieux, C., Fischl, B., Dale, A., & Halgren, E. (2010). Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. NeuroImage, 53, 1–15. https://doi.org/10.1016/j.neuroimage.2010.06.010.
Dhamala, E., Jamison, K.W., Sabuncu, M.R., & Kuceyeski, A. (2020). Sex classification using long-range temporal dependence of resting-state functional MRI time series. Human Brain Mapping, 41, 3567–3579. https://doi.org/10.1002/hbm.25030.
Di Martino, A., Yan, C.-G., Li, Q., Denio, E., Castellanos, F.X., Alaerts, K., & Milham, M.P. (2014). The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry, 19, 659–667. https://doi.org/10.1038/mp.2013.78.
Dong, A., Toledo, J.B., Honnorat, N., Doshi, J., Varol, E., Sotiras, A., & Davatzikos, C. (2017). Heterogeneity of neuroanatomical patterns in prodromal alzheimer’s disease: links to cognition, progression and biomarkers. Brain, aww319. https://doi.org/10.1093/brain/aww319.
Dubois, J., & Adolphs, R. (2016). Building a science of individual differences from fMRI. Trends in Cognitive Sciences, 20, 425–443. https://doi.org/10.1016/j.tics.2016.03.014.
Duchesne, S., Dieumegarde, L., Chouinard, I., Farokhian, F., Badhwar, A., Bellec, P., & Potvin, O. (2019). Structural and functional multi-platform MRI series of a single human volunteer over more than fifteen years. Scientific Data, 6, 245. https://doi.org/10.1038/s41597-019-0262-8.
Dumais, K.M., Chernyak, S., Nickerson, L.D., & Janes, A. C. (2018). Sex differences in default mode and dorsal attention network engagement. PLOS ONE, 13, e0199049. https://doi.org/10.1371/journal.pone.0199049.
DuPre, E., Hanke, M., & Poline, J.-B. (2020). Nature abhors a paywall: How open science can realize the potential of naturalistic stimuli. NeuroImage, 216, 116330. https://doi.org/10.1016/j.neuroimage.2019.116330.
Dwyer, D.B., Falkai, P., & Koutsouleris, N. (2018). Machine learning approaches for clinical psychology and psychiatry. Annual Review of Clinical Psychology, 14, 91–118. https://doi.org/10.1146/annurev-clinpsy-032816-045037.
Easson, A.K., Fatima, Z., & McIntosh, A.R. (2019). Functional connectivity-based subtypes of individuals with and without autism spectrum disorder. Network Neuroscience, 3, 344–362. https://doi.org/10.1162/netn_a_00067.
Eglen, S.J., Marwick, B., Halchenko, Y.O., Hanke, M., Sufi, S., Gleeson, P., & Poline, J.-B. (2017). Toward standard practices for sharing computer code and programs in neuroscience. Nature Neuroscience, 20, 770–773. https://doi.org/10.1038/nn.4550.
Eickhoff, S.B., Yeo, B.T.T., & Genon, S. (2018). Imaging-based parcellations of the human brain. Nature Reviews Neuroscience, 19, 672–686. https://doi.org/10.1038/s41583-018-0071-7.
Ellis, K.A., Bush, A.I., Darby, D., Fazio, D.D., Foster, J., Hudson, P., & And, D.A. (2009). The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. International Psychogeriatrics, 21, 672–687. https://doi.org/10.1017/s1041610209009405.
Engelhardt, L.E., Roe, M.A., Juranek, J., DeMaster, D., Harden, K.P., Tucker-Drob, E.M., & Church, J.A. (2017). Children’s head motion during fMRI tasks is heritable and stable over time. Developmental Cognitive Neuroscience, 25, 58–68. https://doi.org/10.1016/j.dcn.2017.01.011.
Esteban, O., Birman, D., Schaer, M., Koyejo, O.O., Poldrack, R.A., & Gorgolewski, K.J. (2017). MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites. PLOS ONE, 12, e0184661. https://doi.org/10.1371/journal.pone.0184661.
Esteban, O., Markiewicz, C.J., Blair, R.W., Moodie, C.A., Isik, A.I., Erramuzpe, A., & Gorgolewski, K.J. (2019). fMRIPrep: a robust preprocessing pipeline for functional MRI. Nature Methods, 16, 111–116. https://doi.org/10.1038/s41592-018-0235-4.
Fair, D.A., Nigg, J.T., Iyer, S., Bathula, D., Mills, K.L., Dosenbach, N.U.F., & Milham, M.P. (2013). Distinct neural signatures detected for ADHD subtypes after controlling for micro-movements in resting state functional connectivity MRI data. Frontiers in Systems Neuroscience, 6, 80. https://doi.org/10.3389/fnsys.2012.00080.
Fan, L., Li, H., Zhuo, J., Zhang, Y., Wang, J., Chen, L., & Jiang, T. (2016). The Human Brainnetome Atlas: A new brain atlas based on connectional architecture. Cerebral Cortex, 26, 3508–3526. https://doi.org/10.1093/cercor/bhw157.
Ferrucci, L. (2008). The baltimore longitudinal study of aging (BLSA): a 50-year-long journey and plans for the future. Journals of Gerontology Series A: Biological Sciences and Medical Sciences, 63, 1416–1419. https://doi.org/10.1093/gerona/63.12.1416.
Finn, E.S., Glerean, E., Khojandi, A.Y., Nielson, D., Molfese, P.J., Handwerker, D.A., & Bandettini, P.A. (2020). Idiosynchrony: From shared responses to individual differences during naturalistic neuroimaging. NeuroImage, 215, 116828. https://doi.org/10.1016/j.neuroimage.2020.116828.
Finn, E.S., Shen, X., Scheinost, D., Rosenberg, M.D., Huang, J., Chun, M.M., & Constable, R.T. (2015). Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nature Neuroscience, 18, 1664–1671. https://doi.org/10.1038/nn.4135.
Fitzgibbon, S.P., Harrison, S.J., Jenkinson, M., Baxter, L., Robinson, E.C., Bastiani, M., & Andersson, J. (2020). The developing Human Connectome Project (dHCP) automated resting-state functional processing framework for newborn infants. NeuroImage, 223, 117303. https://doi.org/10.1016/j.neuroimage.2020.117303.
Forde, N.J., Jeyachandra, J., Joseph, M., Jacobs, G.R., Dickie, E., Satterthwaite, T.D., & Voineskos, A.N. (2020). Sex differences in variability of brain structure across the lifespan. Cerebral Cortex, 30, 5420–5430. https://doi.org/10.1093/cercor/bhaa123.
Forstmeier, W., & Schielzeth, H. (2011). Cryptic multiple hypotheses testing in linear models: overestimated effect sizes and the winner’s curse. Behavioral Ecology and Sociobiology, 65, 47–55. https://doi.org/10.1007/s00265-010-1038-5.
Frauscher, B., von Ellenrieder, N., Zelmann, R., Doležalová, I., Minotti, L., Olivier, A., & Gotman, J. (2018). Atlas of the normal intracranial electroencephalogram: neurophysiological awake activity in different cortical areas. Brain: A Journal of Neurology, 141, 1130–1144. https://doi.org/10.1093/brain/awy035.
Frazier, J.A., Hodge, S.M., Breeze, J.L., Giuliano, A.J., Terry, J.E., Moore, C.M., & Makris, N. (2008). Diagnostic and sex effects on limbic volumes in early-onset bipolar disorder and schizophrenia. Schizophrenia Bulletin, 34, 37–46. https://doi.org/10.1093/schbul/sbm120.
Froeling, M., Tax, C.M., Vos, S.B., Luijten, P.R., & Leemans, A. (2017). “MASSIVE” brain dataset: Multiple acquisitions for standardization of structural imaging validation and evaluation. Magnetic Resonance in Medicine, 77, 1797–1809. https://doi.org/10.1002/mrm.26259.
Gamss, R.P., Slasky, S.E., Bello, J.A., Miller, T.S., & Shinnar, S. (2009). Prevalence of hippocampal malrotation in a population without seizures. American Journal of Neuroradiology, 30, 1571–1573. https://doi.org/10.3174/ajnr.a1657.
Gan-Or, Z., Rao, T., Leveille, E., Degroot, C., Chouinard, S., Cicchetti, F., & et al. (2020). The Quebec Parkinson Network: A researcher-patient matching platform and multimodal biorepository. Journal of Parkinson’s Disease, 10, 301—313. https://doi.org/10.3233/JPD-191775.
Geerligs, L., & Campbell, K.L. (2018). Age-related differences in information processing during movie watching. Neurobiology of Aging, 72, 106–120. https://doi.org/10.1016/j.neurobiolaging.2018.07.025.
Gilmore, R.O. (2016). From big data to deep insight in developmental science. Wiley Interdisciplinary Reviews: Cognitive Science, 7, 112–126. https://doi.org/10.1002/wcs.1379.
Glasser, M.F., Smith, S.M., Marcus, D.S., Andersson, J.L.R., Auerbach, E.J., Behrens, T.E.J., & Essen, D.C.V. (2016). The Human Connectome Project’s neuroimaging approach. Nature Neuroscience, 19, 1175–1187. https://doi.org/10.1038/nn.4361.
Gollub, R.L., Shoemaker, J.M., King, M.D., White, T., Ehrlich, S., Sponheim, S.R., & Andreasen, N.C. (2013). The MCIC collection: a shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics, 11, 367–388. https://doi.org/10.1007/s12021-013-9184-3.
Gordon, E.M., Laumann, T.O., Gilmore, A.W., Newbold, D.J., Greene, D.J., Berg, J.J., & Dosenbach, N.U.F. (2017). Precision functional mapping of individual human brains. Neuron, 95, 791–807.e7. https://doi.org/10.1016/j.neuron.2017.07.011.
Gorgolewski, K.J., Auer, T., Calhoun, V.D., Craddock, R.C., Das, S., Duff, E.P., & Poldrack, R.A. (2016). The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data, 3. https://doi.org/10.1038/sdata.2016.44.
Gorgolewski, K.J., Mendes, N., Wilfling, D., Wladimirow, E., Gauthier, C.J., Bonnen, T., & Margulies, D.S. (2015). A high resolution 7-Tesla resting-state fMRI test-retest dataset with cognitive and physiological measures. Scientific Data, 2, 140054. https://doi.org/10.1038/sdata.2014.54.
Gorgolewski, K.J., Storkey, A., Bastin, M.E., Whittle, I.R., Wardlaw, J.M., & Pernet, C.R. (2013). A test-retest fMRI dataset for motor, language and spatial attention functions. GigaScience, 2, 6. https://doi.org/10.1186/2047-217x-2-6.
Grady, C.L., Rieck, J.R., Nichol, D., Rodrigue, K.M., & Kennedy, K.M. (2020). Influence of sample size and analytic approach on stability and interpretation of brain-behavior correlations in task-related fMRI data. Human Brain Mapping. https://doi.org/10.1002/hbm.25217.
Grandjean, J., Canella, C., Anckaerts, C., Ayrancı, G., Bougacha, S., Bienert, T., & Gozzi, A. (2020). Common functional networks in the mouse brain revealed by multi-centre resting-state fMRI analysis. NeuroImage, 205, 116278. https://doi.org/10.1016/j.neuroimage.2019.116278.
Gratton, C., Laumann, T.O., Nielsen, A.N., Greene, D.J., Gordon, E.M., Gilmore, A.W., & Petersen, S.E. (2018). Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron, 98, 439–452.e5. https://doi.org/10.1016/j.neuron.2018.03.035.
Gray, J.C., Owens, M.M., Hyatt, C.S., & Miller, J.D. (2018). No evidence for morphometric associations of the amygdala and hippocampus with the five-factor model personality traits in relatively healthy young adults. PLOS ONE, 13, e0204011. https://doi.org/10.1371/journal.pone.0204011.
Greene, A.S., Gao, S., Noble, S., Scheinost, D., & Constable, R. T. (2020). How tasks change whole-brain functional organization to reveal brain-phenotype relationships. Cell Reports, 32, 108066. https://doi.org/10.1016/j.celrep.2020.108066.
Greene, A.S., Gao, S., Scheinost, D., & Constable, R.T. (2018). Task-induced brain state manipulation improves prediction of individual traits. Nature Communications, 9, 2807. https://doi.org/10.1038/s41467-018-04920-3.
Greenland, S. (2019). Valid p-values behave exactly as they should: Some misleading criticisms of p-values and their resolution with s-values. American Statistician, 73, 106–114. https://doi.org/10.1080/00031305.2018.1529625.
Guntupalli, J.S., Feilong, M., & Haxby, J.V. (2018). A computational model of shared fine-scale structure in the human connectome. PLOS Computational Biology, 14, e1006120. https://doi.org/10.1371/journal.pcbi.1006120.
Guntupalli, J.S., Hanke, M., Halchenko, Y.O., Connolly, A.C., Ramadge, P.J., & Haxby, J.V. (2016). A model of representational spaces in human cortex. Cerebral Cortex, 26, 2919–2934. https://doi.org/10.1093/cercor/bhw068.
Guo, S., Xiao, B., & Wu, C. (2020). Identifying subtypes of mild cognitive impairment from healthy aging based on multiple cortical features combined with volumetric measurements of the hippocampal subfields. Quantitative Imaging in Medicine and Surgery, 10, 1477–1489. https://doi.org/10.21037/qims-19-872.
Hagiwara, A., Fujita, S., Ohno, Y., & Aoki, S. (2020). Variability and standardization of quantitative imaging. Investigative Radiology, 55(9), 601–616. https://doi.org/10.1097/rli.0000000000000666.
Hagmann, P., Cammoun, L., Gigandet, X., Meuli, R., Honey, C.J., Wedeen, V.J., & Sporns, O. (2008). Mapping the structural core of human cerebral cortex. PLoS Biology, 6, e159. https://doi.org/10.1371/journal.pbio.0060159.
Hanke, M., Baumgartner, F.J., Ibe, P., Kaule, F.R., Pollmann, S., Speck, O., & Stadler, J. (2014). A high-resolution 7-tesla fMRI dataset from complex natural stimulation with an audio movie. Scientific Data, 1. https://doi.org/10.1038/sdata.2014.3.
Harms, M.P., Somerville, L.H., Ances, B.M., Andersson, J., Barch, D.M., Bastiani, M., & Yacoub, E. (2018). Extending the human connectome project across ages: Imaging protocols for the lifespan development and aging projects. NeuroImage, 183, 972–984. https://doi.org/10.1016/j.neuroimage.2018.09.060.
Hawrylycz, M.J., Lein, E.S., Guillozet-Bongaarts, A.L., Shen, E.H., Ng, L., Miller, J.A., & Jones, A.R. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature, 489(7416), 391–399. https://doi.org/10.1038/nature11405.
Haxby, J.V., Guntupalli, J.S., Connolly, A.C., Halchenko, Y.O., Conroy, B.R., Gobbini, M.I., & Ramadge, P.J. (2011). A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron, 72, 404–416. https://doi.org/10.1016/j.neuron.2011.08.026.
Haxby, J.V., Guntupalli, J.S., Nastase, S.A., & Feilong, M. (2020). Hyperalignment: Modeling shared information encoded in idiosyncratic cortical topographies. eLife, 9, e56601. https://doi.org/10.7554/elife.56601.
Hazlett, H.C., Gu, H., McKinstry, R.C., Shaw, D.W., Botteron, K.N., Dager, S.R., & Network, IBIS. (2012). Brain volume findings in 6-month-old infants at high familial risk for autism. American Journal of Psychiatry, 169, 601–608. https://doi.org/10.1176/appi.ajp.2012.11091425.
Helmer, K.G., Ambite, J.L., Ames, J., Ananthakrishnan, R., Burns, G., Chervenak, A.L., & And, C.K. (2011). Enabling collaborative research using the Biomedical Informatics Research Network (BIRN). Journal of the American Medical Informatics Association, 18, 416–422. https://doi.org/10.1136/amiajnl-2010-000032.
Henson, R.N., Suri, S., Knights, E., Rowe, J.B., Kievit, R.A., Lyall, D.M., & Fisher, S.E. (2020). Effect of apolipoprotein e polymorphism on cognition and brain in the cambridge centre for ageing and neuroscience cohort. Brain and Neuroscience Advances, 4, 239821282096170. https://doi.org/10.1177/2398212820961704.
Herrick, R., Horton, W., Olsen, T., McKay, M., Archie, K.A., & Marcus, D.S. (2016). XNAT Central: Open sourcing imaging research data. NeuroImage, 124, 1093–1096. https://doi.org/10.1016/j.neuroimage.2015.06.076.
Hodge, M.R., Horton, W., Brown, T., Herrick, R., Olsen, T., Hileman, M.E., & Marcus, D.S. (2016). ConnectomeDB–sharing human brain connectivity data. NeuroImage, 124, 1102–1107. https://doi.org/10.1016/j.neuroimage.2015.04.046.
Hodgson, K., Poldrack, R.A., Curran, J.E., Knowles, E.E., Mathias, S., Göring, H. H., & Glahn, D.C. (2017). Shared genetic factors influence head motion during MRI and body mass index. Cerebral Cortex, 27, 5529–5546. https://doi.org/10.1093/cercor/bhw321.
Holmes, A.J., Hollinshead, M.O., O’Keefe, T.M., Petrov, V.I., Fariello, G.R., Wald, L. L., & Buckner, R. L. (2015). Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Scientific Data, 2. https://doi.org/10.1038/sdata.2015.31.
Holmes, A.J., Hollinshead, M.O., Roffman, J.L., Smoller, J.W., & Buckner, R.L. (2016). Individual differences in cognitive control circuit anatomy link sensation seeking, impulsivity, and substance use. Journal of Neuroscience, 36, 4038–4049. https://doi.org/10.1523/jneurosci.3206-15.2016.
Horn, J.D.V., & Gazzaniga, M.S. (2013). Why share data? lessons learned from the fMRIDC. NeuroImage, 82, 677–682. https://doi.org/10.1016/j.neuroimage.2012.11.010.
Horn, J.D.V., Grethe, J.S., Kostelec, P., Woodward, J.B., Aslam, J.A., Rus, D., & Gazzaniga, M.S. (2001). The functional magnetic resonance imaging data center (fMRIDC): the challenges and rewards of large-scale databasing of neuroimaging studies. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 356(1412), 1323–1339. https://doi.org/10.1098/rstb.2001.0916.
Hughes, E.J., Winchman, T., Padormo, F., Teixeira, R., Wurie, J., sharma, M., & hajnal, J.V. (2017). A dedicated neonatal brain imaging system. Magnetic Resonance in Medicine, 78, 794–804. https://doi.org/10.1002/mrm.26462.
Hullett, C.R., & Levine, T.R. (2003). The overestimation of effect sizes from F values in meta-analysis: The cause and a solution. Communication Monographs, 70, 52–67. https://doi.org/10.1080/03637750302475.
Hung, H.M.J., O’Neill, R.T., Bauer, P., & Kohne, K. (1997). The behavior of the p-value when the alternative hypothesis is true. Biometrics, 53, 11. https://doi.org/10.2307/2533093.
Hunt, L.T. (2019). The life-changing magic of sharing your data. Nature Human Behaviour, 3, 312–315. https://doi.org/10.1038/s41562-019-0560-3.
Ince, D.C., Hatton, L., & Graham-Cumming, J. (2012). The case for open computer programs. Nature, 482(7386), 485–488. https://doi.org/10.1038/nature10836.
Jack, C.R., Bernstein, M.A., Fox, N.C., Thompson, P., Alexander, G., Harvey, D., & Weiner, M.W. (2008). The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging, 27, 685–691. https://doi.org/10.1002/jmri.21049.
Jernigan, T.L., Brown, T.T., Hagler, D.J., Akshoomoff, N., Bartsch, H., Newman, E., & Dale, A.M. (2016). The Pediatric Imaging, Neurocognition, and Genetics (PING) data repository. NeuroImage, 124, 1149–1154. https://doi.org/10.1016/j.neuroimage.2015.04.057.
Jiang, R., Zuo, N., Ford, J.M., Qi, S., Zhi, D., Zhuo, C., & Sui, J. (2020). Task-induced brain connectivity promotes the detection of individual differences in brain-behavior relationships. NeuroImage, 207, 116370. https://doi.org/10.1016/j.neuroimage.2019.116370.
Keihaninejad, S., Heckemann, R.A., Gousias, I.S., Rueckert, D., Aljabar, P., Hajnal, J.V., & Hammers, A. (2009). Automatic segmentation of brain MRIs and mapping neuroanatomy across the human lifespan. In Pluim, J. P.W., & Dawant, B. M. (Eds.) Medical imaging 2009: Image processing. https://doi.org/10.1117/12.811429, (Vol. 7259 p. 72591A): SPIE.
Kennedy, D.N., Abraham, S.A., Bates, J.F., Crowley, A., Ghosh, S., Gillespie, T., & Travers, M. (2019). Everything matters: The ReproNim perspective on reproducible neuroimaging. Frontiers in Neuroinformatics, 13, 1. https://doi.org/10.3389/fninf.2019.00001.
Kennedy, D.N., Haselgrove, C., Riehl, J., Preuss, N., & Buccigrossi, R. (2016). The NITRC image repository. NeuroImage, 124, 1069–1073. https://doi.org/10.1016/j.neuroimage.2015.05.074.
Kennedy, K.M., Rodrigue, K.M., Bischof, G.N., Hebrank, A.C., Reuter-Lorenz, P.A., & Park, D.C. (2015). Age trajectories of functional activation under conditions of low and high processing demands: An adult lifespan fMRI study of the aging brain. NeuroImage, 104, 21–34. https://doi.org/10.1016/j.neuroimage.2014.09.056.
Keshavan, A., Datta, E., McDonough, I.M., Madan, C.R., Jordan, K., & Henry, R.G. (2018). Mindcontrol: A web application for brain segmentation quality control. NeuroImage, 170, 365–372. https://doi.org/10.1016/j.neuroimage.2017.03.055.
Kharabian Masouleh, S., Eickhoff, S.B., Zeighami, Y., Lewis, L.B., Dahnke, R., Gaser, C., & Valk, S.L. (2020). Influence of processing pipeline on cortical thickness measurement. Cerebral Cortex, 30, 5014–5027. https://doi.org/10.1093/cercor/bhaa097.
King, R.D., Brown, B., Hwang, M., Jeon, T., & George, A.T. (2010). Fractal dimension analysis of the cortical ribbon in mild Alzheimer’s disease. NeuroImage, 53, 471–479. https://doi.org/10.1016/j.neuroimage.2010.06.050.
King, R.D., George, A.T., Jeon, T., Hynan, L.S., Youn, T.S., Kennedy, D.N., & Dickerson, B. (2009). Characterization of atrophic changes in the cerebral cortex using fractal dimensional analysis. Brain Imaging and Behavior, 3, 154–166. https://doi.org/10.1007/s11682-008-9057-9.
Klapwijk, E.T., van den Bos, W., Tamnes, C.K., Raschle, N.M., & Mills, K.L. (2021). Opportunities for increased reproducibility and replicability of developmental neuroimaging. Developmental Cognitive Neuroscience, 100902. https://doi.org/10.1016/j.dcn.2020.100902.
Klein, A., & Tourville, J. (2012). 101 labeled brain images and a consistent human cortical labeling protocol. Frontiers in Neuroscience, 6, 171. https://doi.org/10.3389/fnins.2012.00171.
Labus, J.S., Naliboff, B., Kilpatrick, L., Liu, C., Ashe-McNalley, C., dos Santos, I.R., & Mayer, E.A. (2016). Pain and Interoception Imaging Network (PAIN): A multimodal, multisite, brain-imaging repository for chronic somatic and visceral pain disorders. NeuroImage, 124, 1232–1237. https://doi.org/10.1016/j.neuroimage.2015.04.018.
Lakens, D., Scheel, A.M., & Isager, P.M. (2018). Equivalence testing for psychological research: A tutorial. Advances in Methods and Practices in Psychological Science, 1, 259–269. https://doi.org/10.1177/2515245918770963.
Lamar, M., Arfanakis, K., Yu, L., Zhang, S., Han, S.D., Fleischman, D.A., & Boyle, P.A. (2020). White matter correlates of scam susceptibility in community-dwelling older adults. Brain Imaging and Behavior, 14, 1521–1530. https://doi.org/10.1007/s11682-019-00079-7.
LaMontagne, P.J., Benzinger, T.L., Morris, J.C., Keefe, S., Hornbeck, R., Xiong, C., & Marcus, D. (2019). OASIS-3: Longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and alzheimer disease. medRxiv. https://doi.org/10.1101/2019.12.13.19014902.
Landis, D., Courtney, W., Dieringer, C., Kelly, R., King, M., Miller, B., & Calhoun, V.D. (2016). COINS Data Exchange: An Open platform for compiling, curating, and disseminating neuroimaging data. NeuroImage, 124, 1084–1088. https://doi.org/10.1016/j.neuroimage.2015.05.049.
Landman, B.A., Huang, A.J., Gifford, A., Vikram, D.S., Lim, I.A.L., Farrell, J.A., & van Zijl, P.C. (2011). Multi-parametric neuroimaging reproducibility: a 3-T resource study. NeuroImage, 54(4), 2854–2866. https://doi.org/10.1016/j.neuroimage.2010.11.047.
Li, G., Zhang, S., Le, T.M., Tang, X., & Li, C.-S. R. (2020). Neural responses to negative facial emotions: Sex differences in the correlates of individual anger and fear traits. NeuroImage, 221, 117171. https://doi.org/10.1016/j.neuroimage.2020.117171.
Li, J., Bolt, T., Bzdok, D., Nomi, J.S., Yeo, B.T.T., Spreng, R.N., & Uddin, L.Q. (2019). Topography and behavioral relevance of the global signal in the human brain. Scientific Reports, 9, 14286. https://doi.org/10.1038/s41598-019-50750-8.
Li, Y., Wang, Z., Boileau, I., Dreher, J.-C., Gelskov, S., Genauck, A., & Sescousse, G. (2019). Altered orbitofrontal sulcogyral patterns in gambling disorder: a multicenter study. Translational Psychiatry, 9, 186. https://doi.org/10.1038/s41398-019-0520-8.
Littlejohns, T.J., Holliday, J., Gibson, L.M., Garratt, S., Oesingmann, N., Alfaro-Almagro, F., & Allen, N.E. (2020). The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nature Communications, 11. https://doi.org/10.1038/s41467-020-15948-9.
Liu, W., Wei, D., Chen, Q., Yang, W., Meng, J., Wu, G., & Qiu, J. (2017). Longitudinal test-retest neuroimaging data from healthy young adults in southwest China. Scientific Data, 4, 170017. https://doi.org/10.1038/sdata.2017.17.
Liu, Y., Perez, P.D., Ma, Z., Ma, Z., Dopfel, D., Cramer, S., & Zhang, N. (2020). An open database of resting-state fMRI in awake rats. NeuroImage, 220, 117094. https://doi.org/10.1016/j.neuroimage.2020.117094.
Longo, D.L., & Drazen, J.M. (2016). Data sharing. New England Journal of Medicine, 374(276), 277. https://doi.org/10.1056/nejme1516564.
Lusebrink, F., Sciarra, A., Mattern, H., Yakupov, R., & Speck, O. (2017). T1-weighted in vivo human whole brain MRI dataset with an ultrahigh isotropic resolution of 250 μ m. Scientific Data, 4, 170032. https://doi.org/10.1038/sdata.2017.32.
Maclaren, J., Han, Z., Vos, S.B., Fischbein, N., & Bammer, R. (2014). Reliability of brain volume measurements: A test-retest dataset. Scientific Data, 1, 140037. https://doi.org/10.1038/sdata.2014.37.
Madan, C.R. (2016). Multiple statistical tests: Lessons from a d20. F1000Research, 5, 1129. https://doi.org/10.12688/f1000research.8834.2.
Madan, C.R. (2017). Advances in studying brain morphology: The benefits of open-access data. Frontiers in Human Neuroscience, 11, 405. https://doi.org/10.3389/fnhum.2017.00405.
Madan, C.R. (2018). Age differences in head motion and estimates of cortical morphology. PeerJ, 6, e5176. https://doi.org/10.7717/peerj.5176.
Madan, C.R. (2019a). Robust estimation of sulcal morphology. Brain Informatics, 6, 5. https://doi.org/10.1186/s40708-019-0098-1.
Madan, C.R. (2019b). Shape-related characteristics of age-related differences in subcortical structures. Aging & Mental Health, 23(800), 810. https://doi.org/10.1080/13607863.2017.1421613.
Madan, C.R. (2021). Age-related decrements in cortical gyrification: Evidence from an accelerated longitudinal dataset. European Journal of Neuroscience, 53, 1661–1671. https://doi.org/10.1111/ejn.15039.
Madan, C.R., & Kensinger, E.A. (2016). Cortical complexity as a measure of age-related brain atrophy. NeuroImage, 134(617), 629. https://doi.org/10.1016/j.neuroimage.2016.04.029.
Madan, C.R., & Kensinger, E.A. (2017a). Age-related differences in the structural complexity of subcortical and ventricular structures. Neurobiology of Aging, 50(87), 95. https://doi.org/10.1016/j.neurobiolaging.2016.10.023.
Madan, C.R., & Kensinger, E.A. (2017b). Test-retest reliability of brain morphology estimates. Brain Informatics, 4, 107–121. https://doi.org/10.1007/s40708-016-0060-4.
Madan, C.R., & Kensinger, E.A. (2018). Predicting age from cortical structure across the lifespan. European Journal of Neuroscience, 47, 399–416. https://doi.org/10.1111/ejn.13835.
Makropoulos, A., Robinson, E.C., Schuh, A., Wright, R., Fitzgibbon, S., Bozek, J., & Rueckert, D. (2018). The developing human connectome project: a minimal processing pipeline for neonatal cortical surface reconstruction. NeuroImage, 173, 88–112. https://doi.org/10.1016/j.neuroimage.2018.01.054.
Mar, R.A., Spreng, R.N., & DeYoung, C.G. (2013). How to produce personality neuroscience research with high statistical power and low additional cost. Cognitive, Affective, & Behavioral Neuroscience, 13, 674–685. https://doi.org/10.3758/s13415-013-0202-6.
Marcus, D.S., Fotenos, A.F., Csernansky, J.G., Morris, J.C., & Buckner, R.L. (2010). Open Access Series of Imaging Studies: Longitudinal MRI data in nondemented and demented older adults. Journal of Cognitive Neuroscience, 22, 2677–2684. https://doi.org/10.1162/jocn.2009.21407.
Marcus, D.S., Harwell, J., Olsen, T., Hodge, M., Glasser, M.F., Prior, F., & Van Essen, D.C. (2011). Informatics and data mining tools and strategies for the human connectome project. Frontiers in Neuroinformatics, 5, 4. https://doi.org/10.3389/fninf.2011.00004.
Marcus, D.S., Olsen, T.R., Ramaratnam, M., & Buckner, R.L. (2007). The extensible neuroimaging archive toolkit. Neuroinformatics, 5, 11–33. https://doi.org/10.1385/ni:5:1:11.
Marcus, D.S., Wang, T.H., Parker, J., Csernansky, J.G., Morris, J.C., & Buckner, R.L. (2007). Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. Journal of Cognitive Neuroscience, 19, 1498–1507. https://doi.org/10.1162/jocn.2007.19.9.1498.
Marek, K., Jennings, D., Lasch, S., Siderowf, A., Tanner, C., Simuni, T., & Taylor, P. (2011). The parkinson progression marker initiative (PPMI). Progress in Neurobiology, 95, 629–635. https://doi.org/10.1016/j.pneurobio.2011.09.005.
Marek, S., Tervo-Clemmens, B., Nielsen, A.N., Wheelock, M.D., Miller, R.L., Laumann, T.O., & Dosenbach, N.U. (2019). Identifying reproducible individual differences in childhood functional brain networks: an ABCD study. Developmental Cognitive Neuroscience, 40, 100706. https://doi.org/10.1016/j.dcn.2019.100706.
Margulies, D.S., Ghosh, S.S., Goulas, A., Falkiewicz, M., Huntenburg, J.M., Langs, G., & Smallwood, J. (2016). Situating the default-mode network along a principal gradient of macroscale cortical organization. Proceedings of the National Academy of Sciences USA, 113, 12574–12579. https://doi.org/10.1073/pnas.1608282113.
Marie, D., Maingault, S., Crivello, F., Mazoyer, B., & Tzourio-Mazoyer, N. (2016). Surface-based morphometry of cortical thickness and surface area associated with heschl’s gyri duplications in 430 healthy volunteers. Frontiers in Human Neuroscience, 10, 69. https://doi.org/10.3389/fnhum.2016.00069.
Marinescu, R.V., Bron, E.E., Oxtoby, N.P., Young, A.L., Toga, A.W., Weiner, M.W., & Alexander, D.C. (2020). Predicting Alzheimer’s disease progression: Results from the TADPOLE challenge. Alzheimer’s & Dementia, 16, S4. https://doi.org/10.1002/alz.039538.
Markett, S., Jawinski, P., Kirsch, P., & Gerchen, M.F. (2020). Specific and segregated changes to the functional connectome evoked by the processing of emotional faces: A task-based connectome study. Scientific Reports, 10, 1. https://doi.org/10.1038/s41598-020-61522-0.
Martino, A.D., O’Connor, D., Chen, B., Alaerts, K., Anderson, J.S., Assaf, M., & Milham, M.P. (2017). Enhancing studies of the connectome in autism using the Autism Brain Imaging Data Exchange II. Scientific Data, 4, 170010. https://doi.org/10.1038/sdata.2017.10.
Mazziotta, J.C., Woods, R., Iacoboni, M., Sicotte, N., Yaden, K., Tran, M., & Toga, A.W. (2009). The myth of the normal, average human brain–the ICBM experience: (1) subject screening and eligibility. NeuroImage, 44, 914–922. https://doi.org/10.1016/j.neuroimage.2008.07.062.
McNabb, C.B., Lindner, M., Shen, S., Burgess, L.G., Murayama, K., & Johnstone, T. (2020). Inter-slice leakage and intra-slice aliasing in simultaneous multi-slice echo-planar images. Brain Structure and Function, 225, 1153–1158. https://doi.org/10.1007/s00429-020-02053-2.
Mendes, N., Oligschlăger, S., Lauckner, M. E, Golchert, J., Huntenburg, J. M., Falkiewicz, M, & Margulies, D. S. (2019). A functional connectome phenotyping dataset including cognitive state and personality measures. Scientific Data, 6, 180307. https://doi.org/10.1038/sdata.2018.307.
Mennes, M., Biswal, B.B., Castellanos, F.X., & Milham, M.P. (2013). Making data sharing work: The FCP/INDI experience. NeuroImage, 82, 683–691. https://doi.org/10.1016/j.neuroimage.2012.10.064.
Milham, M., Petkov, C.I., Margulies, D.S., Schroeder, C.E., Basso, M.A., Belin, P., & di Zhou, Y. (2020). Accelerating the evolution of nonhuman primate neuroimaging. Neuron, 105, 600–603. https://doi.org/10.1016/j.neuron.2019.12.023.
Milham, M.P., Craddock, R.C., Son, J.J., Fleischmann, M., Clucas, J., Xu, H., & Klein, A. (2018). Assessment of the impact of shared brain imaging data on the scientific literature. Nature Communications, 9, 2818. https://doi.org/10.1038/s41467-018-04976-1.
Miller, K.L., Alfaro-Almagro, F., Bangerter, N.K., Thomas, D.L., Yacoub, E., Xu, J., & Smith, S.M. (2016). Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nature Neuroscience, 19, 1523–1536. https://doi.org/10.1038/nn.4393.
Nakamura, M., Nestor, P.G., McCarley, R.W., Levitt, J.J., Hsu, L., Kawashima, T., & Shenton, M.E. (2007). Altered orbitofrontal sulcogyral pattern in schizophrenia. Brain: A Journal of Neurology, 130, 693–707. https://doi.org/10.1093/brain/awm007.
Naselaris, T., Allen, E., & Kay, K. (2021). Extensive sampling for complete models of individual brains. Current Opinion in Behavioral Sciences, 40, 45–51. https://doi.org/10.1016/j.cobeha.2020.12.008.
Nastase, S.A., Liu, Y.-F., Hillman, H., Zadbood, A., Hasenfratz, L., Keshavarzian, N., & Hasson, U. (2020). Narratives: fMRI data for evaluating models of naturalistic language comprehension. bioRxiv, arXiv:2020.12.23.424091. https://doi.org/10.1101/2020.12.23.424091.
Nichols, T.E., Das, S., Eickhoff, S.B., Evans, A.C., Glatard, T., Hanke, M., & Yeo, B.T.T. (2017). Best practices in data analysis and sharing in neuroimaging using MRI. Nature Neuroscience, 20, 299–303. https://doi.org/10.1038/nn.4500.
Nickerson, L.D. (2018). Replication of resting state-task network correspondence and novel findings on brain network activation during task fMRI in the human connectome project study. Scientific Reports, 8, 1. https://doi.org/10.1038/s41598-018-35209-6.
Niso, G., Rogers, C., Moreau, J.T., Chen, L.-Y., Madjar, C., Das, S., & Baillet, S. (2016). OMEGA: The Open MEG archive. NeuroImage, 124, 1182–1187. https://doi.org/10.1016/j.neuroimage.2015.04.028.
Noble, S., Spann, M.N., Tokoglu, F., Shen, X., Constable, R.T., & Scheinost, D. (2017). Influences on the test–retest reliability of functional connectivity MRI and its relationship with behavioral utility. Cerebral Cortex, 27, 5415–5429. https://doi.org/10.1093/cercor/bhx230.
Nooner, K.B., Colcombe, S.J., Tobe, R.H., Mennes, M., Benedict, M.M., Moreno, A.L., & Milham, M.P. (2012). The NKI-rockland Sample: A model for accelerating the pace of discovery science in psychiatry. Frontiers in Neuroscience, 6, 152. https://doi.org/10.3389/fnins.2012.00152.
Nostro, A.D., Müller, V. I., Varikuti, D.P., Plăschke, R. N., Hoffstaedter, F., Langner, R., & Eickhoff, S.B. (2018). Predicting personality from network-based resting-state functional connectivity. Brain Structure and Function, 223, 2699–2719. https://doi.org/10.1007/s00429-018-1651-z.
Orban, P., Madjar, C., Savard, M., Dansereau, C., Tam, A., Das, S., & Bellec, P. (2015). Test-retest resting-state fMRI in healthy elderly persons with a family history of alzheimer’s disease. Scientific Data, 2, 150043. https://doi.org/10.1038/sdata.2015.43.
Owens, M.M., Hyatt, C.S., Gray, J.C., Carter, N.T., MacKillop, J., Miller, J.D., & Sweet, L.H. (2019). Cortical morphometry of the five-factor model of personality: findings from the Human Connectome Project full sample. Social Cognitive and Affective Neuroscience, 14, 381–395. https://doi.org/10.1093/scan/nsz017.
Pardoe, H.R., Hiess, R.K., & Kuzniecky, R. (2016). Motion and morphometry in clinical and nonclinical populations. NeuroImage, 135, 177–185. https://doi.org/10.1016/j.neuroimage.2016.05.005.
Pauli, R., Bowring, A., Reynolds, R., Chen, G., Nichols, T.E., & Maumet, C. (2016). Exploring fMRI results space: 31 variants of an fMRI analysis in AFNI, FSL, and SPM. Frontiers in Neuroinformatics, 10, 24. https://doi.org/10.3389/fninf.2016.00024.
Pernet, C.R., & Madan, C.R. (2020). Data visualization for inference in tomographic brain imaging. European Journal of Neuroscience, 51, 695–705. https://doi.org/10.1111/ejn.14430.
Pierce, H.H., Dev, A., Statham, E., & Bierer, B.E. (2019). Credit data generators for data reuse. Nature, 570(7759), 30–32. https://doi.org/10.1038/d41586-019-01715-4.
Pinho, A.L., Amadon, A., Gauthier, B., Clairis, N., Knops, A., Genon, S., & Thirion, B. (2020). Individual Brain Charting dataset extension, second release of high-resolution fMRI data for cognitive mapping. Scientific Data, 7, 353. https://doi.org/10.1038/s41597-020-00670-4.
Pinho, A.L., Amadon, A., Ruest, T., Fabre, M., Dohmatob, E., Denghien, I., & Thirion, B. (2018). Individual Brain Charting, a high-resolution fMRI dataset for cognitive mapping. Scientific Data, 5. https://doi.org/10.1038/sdata.2018.105.
Podcasy, J.L., & Epperson, C.N. (2016). Considering sex and gender in Alzheimer disease and other dementias. Dialogues in Clinical Neuroscience, 18, 437–446. https://doi.org/10.31887/dcns.2016.18.4.
Poldrack, R.A. (2020). Diving into the deep end: a personal reflection on the MyConnectome study. Current Opinion in Behavioral Sciences, 40, 1–4. https://doi.org/10.1016/j.cobeha.2020.10.008.
Poldrack, R.A., Barch, D.M., Mitchell, J.P., Wager, T.D., Wagner, A.D., Devlin, J.T., & Milham, M.P. (2013). Toward open sharing of task-based fMRI data: the OpenfMRI project. Frontiers in Neuroinformatics, 7, 12. https://doi.org/10.3389/fninf.2013.00012.
Poldrack, R.A., & Gorgolewski, K.J. (2014). Making big data open: Data sharing in neuroimaging. Nature Neuroscience, 17, 1510–1517. https://doi.org/10.1038/nn.3818.
Poldrack, R.A., Congdon, E., Triplett, W., Gorgolewski, K.J., Karlsgodt, K.H., Mumford, J.A., & Bilder, R.M. (2016). A phenome-wide examination of neural and cognitive function. Scientific Data, 3, 160110. https://doi.org/10.1038/sdata.2016.110.
Poldrack, R.A., Baker, C.I., Durnez, J., Gorgolewski, K.J., Matthews, P.M., Munafò, M. R., & Yarkoni, T. (2017). Scanning the horizon: towards transparent and reproducible neuroimaging research. Nature Reviews Neuroscience, 18, 115–126. https://doi.org/10.1038/nrn.2016.167.
Poldrack, R.A., & Gorgolewski, K.J. (2017). OpenfMRI: Open sharing of task fMRI data. NeuroImage, 144, 259–261. https://doi.org/10.1016/j.neuroimage.2015.05.073.
Poldrack, R.A., Gorgolewski, K.J., & Varoquaux, G. (2019). Computational and informatic advances for reproducible data analysis in neuroimaging. Annual Review of Biomedical Data Science, 2, 119–138. https://doi.org/10.1146/annurev-biodatasci-072018-021237.
Poldrack, R.A., Laumann, T.O., Koyejo, O., Gregory, B., Hover, A., Chen, M.-Y., & Mumford, J.A. (2015). Long-term neural and physiological phenotyping of a single human. Nature Communications, 6. https://doi.org/10.1038/ncomms9885.
Power, J.D., Lynch, C.J., Silver, B.M., Dubin, M.J., Martin, A., & Jones, R.M. (2019). Distinctions among real and apparent respiratory motions in human fMRI data. NeuroImage, 201, 116041. https://doi.org/10.1016/j.neuroimage.2019.116041.
Power, J.D., Plitt, M., Laumann, T.O., & Martin, A. (2017). Sources and implications of whole-brain fMRI signals in humans. NeuroImage, 146, 609–625. https://doi.org/10.1016/j.neuroimage.2016.09.038.
Reagh, Z., & Yassa, M. (2017). Selective vulnerabilities and biomarkers in neurocognitive aging. F1000Research, 6, 491. https://doi.org/10.12688/f1000research.10652.1.
Reagh, Z.M., Delarazan, A.I., Garber, A., & Ranganath, C. (2020). Aging alters neural activity at event boundaries in the hippocampus and posterior medial network. Nature Communications, 11, 3980. https://doi.org/10.1038/s41467-020-17713-4.
Riccelli, R., Toschi, N., Nigro, S., Terracciano, A., & Passamonti, L. (2017). Surface-based morphometry reveals the neuroanatomical basis of the five-factor model of personality. Social Cognitive and Affective Neuroscience, 12, 671–684. https://doi.org/10.1093/scan/nsw175.
Richard, G., Kolskår, K., Sanders, A.-M., Kaufmann, T., Petersen, A., Doan, N.T., & Westlye, L.T. (2018). Assessing distinct patterns of cognitive aging using tissue-specific brain age prediction based on diffusion tensor imaging and brain morphometry. PeerJ, 6, e5908. https://doi.org/10.7717/peerj.5908.
Richardson, H., Lisandrelli, G., Riobueno-Naylor, A., & Saxe, R. (2018). Development of the social brain from age three to twelve years. Nature Communications, 9(1), 1027. https://doi.org/10.1038/s41467-018-03399-2.
Risk, B.B., Kociuba, M.C., & Rowe, D.B. (2018). Impacts of simultaneous multislice acquisition on sensitivity and specificity in fMRI. NeuroImage, 172, 538–553. https://doi.org/10.1016/j.neuroimage.2018.01.078.
Romero, J.E., Manjón, J. V., Tohka, J., Coupé, P., & Robles, M. (2015). NABS: non-local automatic brain hemisphere segmentation. Magnetic Resonance Imaging, 33, 474–484. https://doi.org/10.1016/j.mri.2015.02.005.
Salehi, M., Greene, A.S., Karbasi, A., Shen, X., Scheinost, D., & Constable, R.T. (2020). There is no single functional atlas even for a single individual: Functional parcel definitions change with task. NeuroImage, 208, 116366. https://doi.org/10.1016/j.neuroimage.2019.116366.
Satterthwaite, T.D., Connolly, J.J., Ruparel, K., Calkins, M.E., Jackson, C., Elliott, M.A., & Gur, R.E. (2016). The Philadelphia Neurodevelopmental Cohort: A publicly available resource for the study of normal and abnormal brain development in youth. NeuroImage, 124, 1115–1119. https://doi.org/10.1016/j.neuroimage.2015.03.056.
Scheinost, D., Finn, E.S., Tokoglu, F., Shen, X., Papademetris, X., Hampson, M., & Constable, R.T. (2015). Sex differences in normal age trajectories of functional brain networks. Human Brain Mapping, 36(4), 1524–1535. https://doi.org/10.1002/hbm.22720.
Schilling, K.G., Rheault, F., Petit, L, Hansen, C.B, Nath, V, Yeh, F.-C, & Descoteaux, M. (2020). Tractography dissection variability: what happens when 42 groups dissect 14 white matter bundles on the same dataset? bioRxiv, arXiv:2020.10.07.321083. https://doi.org/10.1101/2020.10.07.321083.
Schoffelen, J.-M., Oostenveld, R., Lam, N.H.L., Uddén, J., Hultén, A., & Hagoort, P. (2019). A 204-subject multimodal neuroimaging dataset to study language processing. Scientific Data, 6, 17. Retrieved from https://doi.org/10.1038/s41597-019-0020-y.
Scholtens, L.H., de Reus, M.A., de Lange, S.C., Schmidt, R., & van den Heuvel, M.P. (2018). An MRI von economo-koskinas atlas. NeuroImage, 170, 249–256. https://doi.org/10.1016/j.neuroimage.2016.12.069.
Schönbrodt, F. D., & Perugini, M. (2013). At what sample size do correlations stabilize?. Journal of Research in Personality, 47, 609–612. https://doi.org/10.1016/j.jrp.2013.05.009.
Schrouff, J., Rosa, M.J., Rondina, J.M., Marquand, A.F., Chu, C., Ashburner, J., & Mourão-Miranda, J. (2013). PRONTo: Pattern recognition for neuroimaging toolbox. Neuroinformatics, 11, 319–337. https://doi.org/10.1007/s12021-013-9178-1.
Schultz, D.H., & Cole, M.W. (2016). Higher intelligence is associated with less task-related brain network reconfiguration. Journal of Neuroscience, 36(33), 8551–8561. https://doi.org/10.1523/jneurosci.0358-16.2016.
Schweinsberg, M., Feldman, M., Staub, N., van der Akker, O.R., van Aert, R.C.M., van Assen, M.A.L.M., & Uhlmann, E.L. (2021). Same data, different conclusions: Radical dispersion in empirical results when independent analysts operationalize and test the same hypothesis. Organizational Behavior and Human Decision Processes.
Scott, A., Courtney, W., Wood, D., de la Garza, R., Lane, S., King, M., & Calhoun, V.D. (2011). COINS: An innovative informatics and neuroimaging tool suite built for large heterogeneous datasets. Frontiers in Neuroinformatics, 5, 33. https://doi.org/10.3389/fninf.2011.00033.
Seeliger, K., Sommers, R.P., Güçlü, U., Bosch, S.E., & van Gerven, M.A. J. (2019). A large single-participant fMRI dataset for probing brain responses to naturalistic stimuli in space and time. bioRxiv. https://doi.org/10.1101/687681.
Seitzman, B.A., Gratton, C., Laumann, T.O., Gordon, E.M., Adeyemo, B., Dworetsky, A., & Petersen, S.E. (2019). Trait-like variants in human functional brain networks. Proceedings of the National Academy of Sciences USA, 116, 22851–22861. https://doi.org/10.1073/pnas.1902932116.
Shah, L.M., Cramer, J.A., Ferguson, M.A., Birn, R.M., & Anderson, J.S. (2016). Reliability and reproducibility of individual differences in functional connectivity acquired during task and resting state. Brain and Behavior, 6, e00456. https://doi.org/10.1002/brb3.456.
Shenkin, S.D., Pernet, C., Nichols, T.E., Poline, J.-B., Matthews, P.M., van der Lugt, A., & Wardlaw, J.M. (2017). Improving data availability for brain image biobanking in healthy subjects: Practice-based suggestions from an international multidisciplinary working group. NeuroImage, 153, 399–409. https://doi.org/10.1016/j.neuroimage.2017.02.030.
Shine, J.M., Bissett, P.G., Bell, P.T., Koyejo, O., Balsters, J.H., Gorgolewski, K.J., & Poldrack, R.A. (2016). The dynamics of functional brain networks: Integrated network states during cognitive task performance. Neuron, 92, 544–554. https://doi.org/10.1016/j.neuron.2016.09.018.
Silberzahn, R., Uhlmann, E.L., Martin, D.P., Anselmi, P., Aust, F., Awtrey, E., & Nosek, B.A. (2018). Many analysts, one data set: Making transparent how variations in analytic choices affect results. Advances in Methods and Practices in Psychological Science, 1, 337–356. https://doi.org/10.1177/2515245917747646.
Smith, S.M., Nichols, T.E., Vidaurre, D., Winkler, A.M., Behrens, T.E.J., Glasser, M.F., & Miller, K.L. (2015). A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nature Neuroscience, 18, 1565–1567. https://doi.org/10.1038/nn.4125.
Snoek, L., van der Miesen, M.M., Beemsterboer, T., van der Leij, A., Eigenhuis, A., & Scholte, H.S. (2021). The Amsterdam Open MRI Collection, a set of multimodal MRI datasets for individual difference analyses. Scientific Data, 8, 85. https://doi.org/10.1038/s41597-021-00870-61.
Somerville, L.H., Bookheimer, S.Y., Buckner, R.L., Burgess, G.C., Curtiss, S.W., Dapretto, M., & Barch, D.M. (2018). The Lifespan Human Connectome Project in Development: A large-scale study of brain connectivity development in 5–21 year olds. NeuroImage, 183, 456–468. https://doi.org/10.1016/j.neuroimage.2018.08.050.
Souza, R., Lucena, O., Garrafa, J., Gobbi, D., Saluzzi, M., Appenzeller, S., & Lotufo, R. (2018). An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. NeuroImage, 170, 482–494. https://doi.org/10.1016/j.neuroimage.2017.08.021.
Spronk, M., Keane, B.P., Ito, T., Kulkarni, K., Ji, J.L., Anticevic, A., & Cole, M.W. (2021). A whole-brain and cross-diagnostic perspective on functional brain network dysfunction. Cerebral Cortex, 31, 547–561. https://doi.org/10.1093/cercor/bhaa242.
Sripada, C., Angstadt, M., Rutherford, S., Kessler, D., Kim, Y., Yee, M., & Levina, E. (1900). Basic units of inter-individual variation in resting state connectomes. Scientific Reports, 9. https://doi.org/10.1038/s41598-018-38406-5.
Steegen, S., Tuerlinckx, F., Gelman, A., & Vanpaemel, W. (2016). Increasing transparency through a multiverse analysis. Perspectives on Psychological Science, 11, 702–712. https://doi.org/10.1177/1745691616658637.
Stephen, J.M., Solis, I., Janowich, J., Stern, M., Frenzel, M.R., Eastman, J.A., & Calhoun, V.D. (2021). The Developmental Chronnecto-Genomics (dev-CoG) study: A multimodal study on the developing brain. NeuroImage, 225, 117438. https://doi.org/10.1016/j.neuroimage.2020.117438.
Tabrizi, S.J., Langbehn, D.R., Leavitt, B.R., Roos, R.A., Durr, A., Craufurd, D., & Stout, J.C. (2009). Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurology, 8, 791–801. https://doi.org/10.1016/s1474-4422(09)70170-x.
Tang, S., Sun, N., Floris, D.L., Zhang, X., Martino, A.D., & Yeo, B.T. (2020). Reconciling dimensional and categorical models of autism heterogeneity: A brain connectomics and behavioral study. Biological Psychiatry, 87, 1071–1082. https://doi.org/10.1016/j.biopsych.2019.11.009.
Taylor, A.M., Pattie, A., & Deary, I.J. (2018). Cohort Profile Update: The Lothian Birth Cohorts of 1921 and 1936. International Journal of Epidemiology, 47(4), 1042–1042r. https://doi.org/10.1093/ije/dyy022.
Taylor, J.R., Williams, N., Cusack, R., Auer, T., Shafto, M.A., Dixon, M., & Henson, R.N. (2017). The Cambridge Centre for Ageing and Neuroscience (cam-CAN) data repository: Structural and functional MRI, MEG, and cognitive data from a cross-sectional adult lifespan sample. NeuroImage, 144, 262–269. https://doi.org/10.1016/j.neuroimage.2015.09.018.
Thiebaut de Schotten, M., Dell’Acqua, F., Ratiu, P., Leslie, A., Howells, H., Cabanis, E., & Catani, M. (2015). From Phineas Gage and Monsieur Leborgne to H.M.: Revisiting disconnection syndromes. Cerebral Cortex, 25, 4812–4827. https://doi.org/10.1093/cercor/bhv173.
Termenon, M., Jaillard, A., Delon-Martin, C., & Achard, S. (2016). Reliability of graph analysis of resting state fMRI using test-retest dataset from the human connectome project. NeuroImage, 142, 172–187. https://doi.org/10.1016/j.neuroimage.2016.05.062.
Thiese, M.S., Ronna, B., & Ott, U. (2016). P value interpretations and considerations. Journal of Thoracic Disease, 8, E928–E931. https://doi.org/10.21037/jtd.2016.08.16.
Thompson, W.H., Wright, J., Bissett, P.G., & Poldrack, R.A. (2020). Dataset decay and the problem of sequential analyses on open datasets. eLife, 9, 53498. https://doi.org/10.7554/elife.53498.
Todd, N., Moeller, S., Auerbach, E.J., Yacoub, E., Flandin, G., & Weiskopf, N. (2016). Evaluation of 2d multiband EPI imaging for high-resolution, whole-brain, task-based fMRI studies at 3 T: Sensitivity and slice leakage artifacts. NeuroImage, 124, 32–42. https://doi.org/10.1016/j.neuroimage.2015.08.056.
Tremblay-Mercier, J., Madjar, C., Das, S., Dyke, S.O., Étienne, P., Lafaille-Magnan, M.-E., & PREVENT-AD Research Group. (2020). Creation of an open science dataset from PREVENT-AD, a longitudinal cohort study of pre-symptomatic Alzheimer’s disease. bioRxiv, arXiv:2020.03.04.976670. https://doi.org/10.1101/2020.03.04.976670.
Tustison, N.J., Cook, P.A., Klein, A., Song, G., Das, S.R., Duda, J.T., & Avants, B.B. (2014). Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. NeuroImage, 99, 166–179. https://doi.org/10.1016/j.neuroimage.2014.05.044.
Uddin, L.Q., Yeo, B.T.T., & Spreng, R.N. (2019). Towards a universal taxonomy of macro-scale functional human brain networks. Brain Topography, 32, 926–942. https://doi.org/10.1007/s10548-019-00744-6.
Valdes-Sosa, P.A., Galan-Garcia, L., Bosch-Bayard, J., Bringas-Vega, M.L., Aubert-Vazquez, E., Rodriguez-Gil, I., & Valdes-Sosa, M.J. (2021). The Cuban Human Brain Mapping Project, a young and middle age population-based EEG, MRI, and cognition dataset. Scientific Data, 8, 45. https://doi.org/10.1038/s41597-021-00829-7.
van der Linden, D., Dunkel, C.S., & Madison, G. (2017). Sex differences in brain size and general intelligence (g ). Intelligence, 63, 78–88. https://doi.org/10.1016/j.intell.2017.04.007.
van Eijk, L., Hansell, N.K., Strike, L.T., Couvy-Duchesne, B., de Zubicaray, G.I., Thompson, P.M., & Wright, M.J. (2020). Region-specific sex differences in the hippocampus. NeuroImage, 215, 116781. https://doi.org/10.1016/j.neuroimage.2020.116781.
Van Essen, D.C., Smith, S.M., Barch, D.M., Behrens, T.E.J., Yacoub, E., & Ugurbil, K. (2013). The WU-Minn Human Connectome Project: An overview. NeuroImage, 80, 62–79. https://doi.org/10.1016/j.neuroimage.2013.05.041.
Vanderwal, T., Eilbott, J., & Castellanos, F.X. (2019). Movies in the magnet: Naturalistic paradigms in developmental functional neuroimaging. Developmental Cognitive Neuroscience, 36, 100600. https://doi.org/10.1016/j.dcn.2018.10.004.
Varoquaux, G. (2018). Cross-validation failure: Small sample sizes lead to large error bars. NeuroImage, 180, 68–77. https://doi.org/10.1016/j.neuroimage.2017.06.061.
Vogt, B.A., Nimchinsky, E.A., Vogt, L.J., & Hof, P.R. (1995). Human cingulate cortex: Surface features, flat maps, and cytoarchitecture. Journal of Comparative Neurology, 359(3), 490–506. https://doi.org/10.1002/cne.903590310.
Vul, E., Harris, C., Winkielman, P., & Pashler, H. (2009). Puzzlingly high correlations in fMRI studies of emotion, personality, and social cognition. Perspectives on Psychological Science, 4, 274–290. https://doi.org/10.1111/j.1745-6924.2009.01125.x.
Wang, L., Alpert, K.I., Calhoun, V.D., Cobia, D.J., Keator, D.B., King, M.D., & Ambite, J.L. (2016). Schizconnect: Mediating neuroimaging databases on schizophrenia and related disorders for large-scale integration. NeuroImage, 124, 1155–1167. https://doi.org/10.1016/j.neuroimage.2015.06.065.
Wang, L., Kogan, A., Cobia, D., Alpert, K., Kolasny, A., Miller, M.I., & Marcus, D. (2013). Northwestern University Schizophrenia Data and Software Tool (NUSDAST). Frontiers in Neuroinformatics, 7, 25. https://doi.org/10.3389/fninf.2013.00025.
Wang, Y., Necus, J., Kaiser, M., & Mota, B. (2016). Universality in human cortical folding in health and disease. Proceedings of the National Academy of Sciences USA, 113, 12820–12825. https://doi.org/10.1073/pnas.1610175113.
Wei, D., Zhuang, K., Ai, L., Chen, Q., Yang, W., Liu, W., & Qiu, J. (2018). Structural and functional brain scans from the cross-sectional Southwest University adult lifespan dataset. Scientific Data, 5, 180134. https://doi.org/10.1038/sdata.2018.134.
Weidemann, C.T., Kragel, J.E., Lega, B.C., Worrell, G.A., Sperling, M.R., Sharan, A.D., & Kahana, M.J. (2019). Neural activity reveals interactions between episodic and semantic memory systems during retrieval. Journal of Experimental Psychology: General, 148, 1–12. https://doi.org/10.1037/xge0000480.
Weiss, T., Soroka, T., Gorodisky, L., Shushan, S., Snitz, K., Weissgross, R., & Sobel, N. (2020). Human olfaction without apparent olfactory bulbs. Neuron, 105, 35–45.e5. https://doi.org/10.1016/j.neuron.2019.10.006.
Westfall, J., Nichols, T.E., & Yarkoni, T. (2017). Fixing the stimulus-as-fixed-effect fallacy in task fMRI. Wellcome Open Research, 1, 23. https://doi.org/10.12688/wellcomeopenres.10298.2.
Weston, S.J., Ritchie, S.J., Rohrer, J.M., & Przybylski, A.K. (2019). Recommendations for increasing the transparency of analysis of preexisting data sets. Advances in Methods and Practices in Psychological Science, 2, 214–227. https://doi.org/10.1177/2515245919848684.
White, T., Blok, E., & Calhoun, V.D. (2020). Data sharing and privacy issues in neuroimaging research: Opportunities, obstacles, challenges, and monsters under the bed. Human Brain Mapping. https://doi.org/10.1002/hbm.25120.
Wierenga, L.M., Sexton, J.A., Laake, P., Giedd, J.N., Tamnes, C.K., & The Pediatric Imaging, Neurocognition, and Genetics Study. (2018). A key characteristic of sex differences in the developing brain: Greater variability in brain structure of boys than girls. Cerebral Cortex, 28, 2741–2751. https://doi.org/10.1093/cercor/bhx154.
Wilkinson, M.D., Dumontier, M., Aalbersberg, I.J., Appleton, G., Axton, M., Baak, A., & Mons, B. (2016). The FAIR guiding principles for scientific data management and stewardship. Scientific Data, 3, 160018. https://doi.org/10.1038/sdata.2016.18.
Wilson, G., Bryan, J., Cranston, K., Kitzes, J., Nederbragt, L., & Teal, T.K. (2017). Good enough practices in scientific computing. PLOS Computational Biology, 13, e1005510. https://doi.org/10.1371/journal.pcbi.1005510.
Yamashita, A., Yahata, N., Itahashi, T., Lisi, G., Yamada, T., Ichikawa, N., & Imamizu, H. (2019). Harmonization of resting-state functional MRI data across multiple imaging sites via the separation of site differences into sampling bias and measurement bias. PLOS Biology, 17, e3000042. https://doi.org/10.1371/journal.pbio.3000042.
Yang, G., Bozek, J., Han, M., & Gao, J.-H. (2020). Constructing and evaluating a cortical surface atlas and analyzing cortical sex differences in young chinese adults. Human Brain Mapping, 41, 2495–2513. https://doi.org/10.1002/hbm.24960.
Yang, H.-C.S., Liang, Z., Yao, J.F., Shen, X., Frederick, D.E.B.B., & Tong, Y. (2018). Vascular effects of caffeine found in BOLD fMRI. Journal of Neuroscience Research, 97, 456–466. https://doi.org/10.1002/jnr.24360.
Ye, B., Khan, S.S., Chikhaoui, B., Iaboni, A., Martin, L.S., Newman, K., & Mihailidis, A. (2019). Challenges in collecting big data in a clinical environment with vulnerable population: Lessons learned from a study using a multi-modal sensors platform. Science and Engineering Ethics, 25, 1447–1466. https://doi.org/10.1007/s11948-018-0072-y.
Yeo, B.T., Krienen, F.M., Chee, M.W., & Buckner, R.L. (2014). Estimates of segregation and overlap of functional connectivity networks in the human cerebral cortex. NeuroImage, 88, 212–227. https://doi.org/10.1016/j.neuroimage.2013.10.046.
Young, A.L., Marinescu, R.V., Oxtoby, N.P., Bocchetta, M., Yong, K., Firth, N.C., & Alexander, D.C. (2018). Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with subtype and stage inference. Nature Communications, 9, 4273. https://doi.org/10.1038/s41467-018-05892-0.
Young, J.T., Shi, Y., Niethammer, M., Grauer, M., Coe, C.L., Lubach, G.R., & Styner, M.A. (2017). The UNC-wisconsin Rhesus Macaque Neurodevelopment Database: A structural MRI and DTI database of early postnatal development. Frontiers in Neuroscience, 11, 29. https://doi.org/10.3389/fnins.2017.00029.
Yun, H.J., Im, K., Yang, J.-J., Yoon, U., & Lee, J.-M. (2013). Automated sulcal depth measurement on cortical surface reflecting geometrical properties of sulci. PLoS ONE, 8, e55977. https://doi.org/10.1371/journal.pone.0055977.
Zacà, D., Hasson, U., Minati, L., & Jovicich, J. (2018). Method for retrospective estimation of natural head movement during structural MRI. Journal of Magnetic Resonance Imaging, 48, 927–937. https://doi.org/10.1002/jmri.25959.
Zhang, X., Mormino, E.C., Sun, N., Sperling, R.A., Sabuncu, M.R., & Yeo, B.T.T. (2016). Bayesian model reveals latent atrophy factors with dissociable cognitive trajectories in Alzheimer’s disease. Proceedings of the National Academy of Sciences USA, 113, E6535–E6544. https://doi.org/10.1073/pnas.1611073113.
Zuo, X.-N., Anderson, J.S., Bellec, P., Birn, R.M., Biswal, B.B., Blautzik, J., & Milham, M.P. (2014). An open science resource for establishing reliability and reproducibility in functional connectomics. Scientific Data, 1, 140049. https://doi.org/10.1038/sdata.2014.49.
Zuo, N., Yang, Z., Liu, Y., Li, J., & Jiang, T. (2017). Both activated and less-activated regions identified by functional MRI reconfigure to support task executions. Brain and Behavior, 8, e00893. https://doi.org/10.1002/brb3.893.
Zuo, X.-N., Xu, T., & Milham, M.P. (2019). Harnessing reliability for neuroscience research. Nature Human Behaviour, 3, 768–771. https://doi.org/10.1038/s41562-019-0655-x.
Acknowledgments
An earlier form of this article was prepared as supplementary to a workshop of the same name, for the fourth Federation of the European Societies of Neuropsychology (FESN) scientific school in August 2020.
I would like to thank Simon Eickhoff for insightful discussions, as well as Andrew Reid for comments on an earlier draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Information Sharing Statement
No new data or software were created in relation to this article.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Madan, C.R. Scan Once, Analyse Many: Using Large Open-Access Neuroimaging Datasets to Understand the Brain. Neuroinform 20, 109–137 (2022). https://doi.org/10.1007/s12021-021-09519-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-021-09519-6