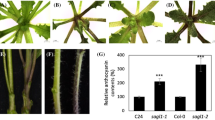
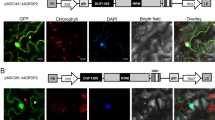
Abstract
The F-box domain is a conserved structural protein motif that most frequently interacts with the SKP1 protein, the core of the SCFs (SKP1-CULLIN-F-box protein ligase) E3 ubiquitin protein ligases. As part of the SCF complexes, the various F-box proteins recruit substrates for degradation through ubiquitination. In this study, we functionally characterized an F-box gene (MtF-box) identified earlier in a population of Tnt1 retrotransposon-tagged mutants of Medicago truncatula and its Arabidopsis thaliana homolog (AtF-box) using gain- and loss-of-function plants. We highlighted the importance of MtF-box in leaf development of M. truncatula. Protein–protein interaction analyses revealed the 2-isopropylmalate synthase (IPMS) protein as a common interactor partner of MtF-box and AtF-box, being a key enzyme in the biosynthesis pathway of the branched-chain amino acid leucine. For further detailed analysis, we focused on AtF-box and its role during the cell division cycle. Based on this work, we suggest a mechanism for the role of the studied F-box gene in regulation of leucine homeostasis, which is important for growth.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this manuscript [and its electronic supplementary material]. The data and the plant material will be available upon request.
References
Abràmoff MD, Magelhaes PJ, Ram SJ (2004) Image Processing with ImageJ. Biophotonics Intern 11:36–42
Baute J, Polyn S, De Block J, Blomme J, Van Lijsebettens M, Inzé D (2017) F-box protein FBX92 affects leaf size in Arabidopsis thaliana. Plant Cell Physiol 58:962–975
Benderoth M, Pfalz M, Kroymann J (2009) Methylthioalkylmalate synthases: genetics, ecology and evolution. Phytochem Rev 8:255–268
Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E (2010) Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22:1264–1280
Boycheva I, Vassileva V, Revalska M, Zehirov G, Iantcheva A (2015) Cyclin-like F-box protein plays a role in growth and development of the three model species Medicago truncatula, Lotus japonicus, and Arabidopsis thaliana. Res Rep Biol 6:117–130
Bu Q, Lv T, Shen H, Phi L, Wang J, Wang Z, Huang Z, Xiao L, Engineer C, Kim TH et al (2014) Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol 164:424–439
Bürckstümmer T, Bennett KL, Preradovic A, Schütze G, Hantschel O, Superti-Furga G, Bauch A (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3:1013–1019
Cao P, Kim S-J, Xing A, Schenck CA, Liu L, Jiang N, Wang J, Last RL, Brandizzi F (2019) Homeostasis of branched-chain amino acids is critical for the activity of TOR signaling in Arabidopsis. eLife 8:e50747. https://doi.org/10.7554/eLife.50747
Cano-Crespo S, Chillarón J, Junza A, Fernández-Miranda G, García J, Polte C, de la Ballina LR, Ignatova Z, Yanes Ó, Zorzano A, Attolini CS-O, Palacín M (2019) CD98hc (SLC3A2) sustains amino acid and nucleotide availability for cell cycle progression. Sci Rep 9:14065. https://doi.org/10.1038/s41598-019-50547-9
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cools T, Iantcheva A, Maes S, Van den Daele H, De Veylder L (2010) A replication stress-induced synchronization method for Arabidopsis thaliana root meristems. Plant J 64:705–714
d’Erfurth I, Cosson V, Eschstruth A, Lucas H, Kondorosi A, Ratet P (2003) Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J 34:95–106
de Kraker J-W, Luck K, Textor S, Tokuhisa JG, Gershenzon J (2007) Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol 143:970–986
del Pozo CJ, Estelle M (2000) F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol Biol 44:123–128
del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell 14:3057–3071
del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin SCFSKP2A pathway in Arabidopsis. Plant Cell 18:2224–2235
del Pozo JC, Manzano C (2013) Auxin and the ubiquitin pathway. Two players-one target: the cell cycle in action. J Exp Bot 65:2617–2632
Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39:W13–W17
De Veylder L, Beeckman T, Beemster GTS, de Almeida EJ, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inzé D (2002) Control of proliferation endoreduplication and differentiation by the Arabidopsis E2Fa/DPa transcription factor. EMBO J 21:1360–1368
Gonzalez-Carranza ZH, Rompa U, Peters JL, Bhatt AM, Wagstaff C, Stead AD, Roberts JA (2007) Hawaiian skirt: an F-box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol 144:1370–1382
Goossens J, Geyter De, Walton A, Eeckhout D, Mertens J, Pollier J, Fiallos-Jurado J, De Keyser A, De Clercq R, Van Leene J, Gevaert K, De Jaeger G, Goormachtig S, Goossens A (2016) Isolation of protein complexes from the model legume Medicago truncatula by tandem affinity purification in hairy root cultures. Plant J 88:476–489
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Gusti A, Baumberger N, Nowack M, Pusch S, Eisler H, Potuschak T, De Veylder L, Schnittger A, Genschik P (2009) The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS ONE 4(3):e4780. https://doi.org/10.1371/journal.pone.0004780
Hummel J, Strehmel N, Selbig J, Walther D, Kopka J (2010) Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 6:322–333
Iantcheva A, Boycheva I, Revalska M (2015) Development of root tips synchronized system for the model legume Medicago truncatula upon replication stress. Bulg J Agric Sci 21:1177–1184
Iantcheva A, Chabaud M, Cosson V, Barascud M, Schutz B, Primard-Brisset C, Durand P, Barker DG, Vlahova M, Ratet P (2009) Osmotic shock improves Tnt1 transposition frequency in Medicago truncatula cv. Jemalong during in vitro regeneration. Plant Cell Rep 28:1563–1572
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Jia FJ, Wu BJ, Li H, Huang JG, Zheng CC (2013) Genome-wide identification and characterisation of F-box family in maize. Mol Genet Genomics 288:559–577
Jia Q, Xiao Z-X, Wong F-L, Sun S, Liang K-J, Lam H-M (2017) Genome-wide analyses of the soybean F-box gene family in response to salt stress. Int J Mol Sci 18:E818. https://doi.org/10.3390/ijms18040818
Jurado S, Abraham Z, Manzano C, Lopez-Torrejon G, Pacios LF, del Pozo JC (2010) The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22:3891–3904
Karimi M, Bleys A, Vanderhaeghen R, Hilson P (2007) Building blocks for plant gene assembly. Plant Physiol 145:1183–1191
Kim HJ, Oh SA, Brownfield L, Hong SH, Ryu H, Hwang I, Twell D, Nam HG (2008) Control of plant germline proliferation by SCFFBL17 degradation of cell cycle inhibitors. Nature 455:1134–1137
Kuroda H, Yanagawa Y, Takahashi N, Horii Y, Matsui M (2012) A comprehensive analysis of interaction and localization of Arabidopsis SKP1-like (ASK) and F-box (FBX) proteins. PLoS ONE 7:e50009. https://doi.org/10.1371/journal.pone.0050009
Li Y, Zhang L, Li D, Liu Z, Wang J, Li X, Yang Y (2016) The Arabidopsis F-box E3 ligase RIFP1 plays a negative role in abscisic acid signalling by facilitating ABA receptor RCAR3 degradation. Plant Cell Environ 39:571–582
Majee M, Kumar S, Kathare PK, Wu S, Gingerich D, Nayak NR, Salaita L, Dinkins R, Martin K, Goodin M, Dirk LMA, Lloyd TD, Zhu L, Chappell J, Hunt AG, Vierstra R, Huq E, Downie AB (2018) KELCH F-BOX protein positively influences Arabidopsis seed germination by targeting PHYTOCHROMEINTERACTING FACTOR1. Proc Natl Acad Sci USA 115:E4120-4129. https://doi.org/10.1073/pnas.1711919115
Manion RE, Huie RD, Levin DR, Manion JA, Huie RE, Levin RD, Burgess Jr. DR, Orkin VL, Tsang W, McGivern WS, Hudgens JW, Knyazev VD, Atkinson DB, Chai E, Tereza AM, Lin C-Y, Allison TC, Mallard WG, Westley F, Herron JT, Hampson RF, Frizzell DH (2015) NIST Chemical Kinetics Database, NIST Standard Reference Database 17, Version 7.0 (Web Version), Release 1.6.8, Data version 2015.09, National Institute of Standards and Technology, Gaithersburg, Maryland, 20899–208320. http://kinetics.nist.gov/
Menges M, Hennig L, Gruissem W, Murray JA (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53:423–442
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nolan KE, Rose RJ, Gorst JG (1989) Regeneration of Medicago truncatula from tissue culture: increased somatic embryogenesis using explant from regenerated plants. Plant Cell Rep 8:278–281
Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for multiple sequence alignments. J Mol Biol 302:205–217
Obayashi T, Aoki Y, Tadaka S, Kagaya Y, Kinoshita K (2018) ATTED-II in 2018: a plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol 59:e3. https://doi.org/10.1093/pcp/pcx191
Revalska M, Vassileva V, Goormachtig S, Van Hautegem T, Ratet P, Ianctheva A (2011) Recent progress in development of a Tnt1 functional genomics platform for the model legumes Medicago truncatula and Lotus japonicus in Bulgaria. Curr Genomics 12:147–152
Revalska M, Vassileva V, Zechirov G, Iantcheva A (2015) Is the auxin influx carrier LAX3 essential for plant growth and development in the model plants Medicago truncatula, Lotus japonicus and Arabidopsis thaliana? Biotechnol Biotechnol Equip 29:786–797
Revalska M, Vassileva V, Zehirov G, Goormachtig S, Iantcheva A (2017) Assessment of the function and expression pattern of auxin response factor B3 in the model legume plant Medicago truncatula. Turk J Biol 41:66–76
Roitinger E, Hofer M, Köcher T, Pichler P, Novatchkova M, Yang J, Schlögelhofer P, Mechtler K (2015) Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol Cell Proteomics 14:556–571
Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336:1045–1049
Song JB, Wang YX, Li HB, Li BW, Zhou ZS, Gao S, Yang ZM (2015) The F-box family genes as key elements in response to salt, heavy mental, and drought stresses in Medicago truncatula. Funct Integr Genomics 15:495–507
Stone SL, Callis J (2007) Ubiquitin ligases mediate growth and development by promoting protein death. Curr Opin Plant Biol 10:624–632
Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K (2017) PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res 46:D1190–D1196. https://doi.org/10.1093/nar/gkx1002
Vandepoele K, Van Bel M, Richard G, Van Landeghem S, Verhelst B, Moreau H, Van de Peer Y, Grimsley N, Piganeau G (2013) pico-PLAZA, a genome database of microbial photosynthetic eukaryotes. Environ Microbiol 15:2147–2153
Van Leene J, Stals H, Eeckhout D, Persiau G, van De Slijke E, van Isterdael G, De Clercq A, Bonnet E, Laukens K, Remmerie N, Henderickx K, De Vijlder T, Abdelkrim A, Pharazyn A, van Onckelen H, Inze D, Witters E, Jaeger De (2007) A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics 6:1226–1238
Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, De Bodt S, Maere S, Laukens K, Pharazyn A, Ferreira PC, Eloy N, Renne C, Meyer C, Faure JD, Steinbrenner J, Beynon J, Larkin JC, Van de Peer Y, Hilson P, Kuiper M, De Veylder L, Van Onckelen H, Inzé D, Witters E, De Jaeger G (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6:397. https://doi.org/10.1038/msb.2010.53
Van Leene J, Witters E, Inzé D, De Jaeger G (2008) Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci 13:517–520
Vierstra RD (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10:385–397
Waese J, Provart NJ (2016) The Bio-Analytic Resource for Plant Biology. Book: eds. Aalt DJ van Dijk, Plant Genomics Databases: Methods and Protocols, Methods Mol Biol 1533:119–148
Walton A, Stes E, Cybulski N, Van Bel M, Iñigo S, Durand AN, Timmerman E, Heyman J, Pauwels L, De Veylder L, Goossens A, De Smet I, Coppens F, Goormachtig S, Gevaert K (2016) It’s time for some “site”-seeing: novel tools to monitor the ubiquitin landscape in Arabidopsis thaliana. Plant Cell 28:6–16
Xu G, Ma H, Neic M, Kong H (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA 106:835–840
Young ND, Udvardi M (2009) Translating Medicago truncatula genomics to crop legumes. Curr Opin Plant Biol 12:193–201
Yu H, Zhang F, Wang G, Liu Y, Liu D (2013) Partial deficiency of isoleucine impairs root development and alters transcript levels of the genes involved in branched-chain amino acid and glucosinolate metabolism in Arabidopsis. J Exp Bot 64:599–612
Zhang X, Abrahan C, Colquhoun TA, Liu C-J (2017) A proteolytic regulator controlling Chalcone synthase stability and flavonoid biosynthesis in Arabidopsis. Plant Cell 29:1157–1174
Zhang X, Gou M, Liu C-J (2013) Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25:4994–5010
Zhao X, Harashima H, Dissmeyer N, Pusch S, Weimer AK, Bramsiepe J, Bouyer D, Rademacher S, Nowack MK, Novak B, Sprunck S, Schnittger A (2012) A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet 8:e1002847. https://doi.org/10.1371/journal.pgen.1002847
Zhao Z, Zhang G, Zhou S, Ren Y, Wang W (2017) The improvement of salt tolerance in transgenic tobacco by overexpression of wheat F-box gene TaFBA1. Plant Sci 259:71–85
Acknowledgements
Part of the research was supported by EMBO fellowship (ASTF 506-2011) to Dr. Anelia Iantcheva, carried out at the Department of Plant Systems Biology, VIB, Ghent University, Ghent, Belgium, supervised by Prof. Lieven De Veylder. The authors wish to thank Dr. Valya Vassileva and Dr. Grigor Zehirov (Institute of Plant Physiology and Genetics, Bulgarian Academy of Sciences) for the establishment and maintenance of hydroponic culture of M. truncatula plants and technical help of Kety Krastanova (Agrobioinstitute).
Funding
Part of the research was supported by EMBO fellowship (ASTF 506–2011) to Dr. Anelia Iantcheva, carried out at the Department of Plant Systems Biology, VIB, Ghent University, Ghent, Belgium, supervised by Prof. Lieven De Veylder.
Author information
Authors and Affiliations
Contributions
AI performed most of the experiments and data analyses. AI and MZ wrote the manuscript, designed and prepared the figures. MR performed phenotyping of A. thaliana transgenic lines, IB contributed with sample preparation for GC–MS of M. truncatula, JH performed F-box cell cycle localization and contributed for A. thaliana TAP analysis, IlB and ID performed the GC–MS analyses, NG was responsible for proteomics data of M. truncatula, SG was responsible for the experimental design of M. truncatula TAP data analyses, LV advised on experimental design and revised the paper. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All the authors have approved their participation in the final manuscript.
Consent for publication
All the authors have read and approved the final manuscript and its submission for publication.
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Anne-Catherine Schmit
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Iantcheva, A., Zhiponova, M., Revalska, M. et al. A common F-box gene regulates the leucine homeostasis of Medicago truncatula and Arabidopsis thaliana. Protoplasma 259, 277–290 (2022). https://doi.org/10.1007/s00709-021-01662-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-021-01662-w