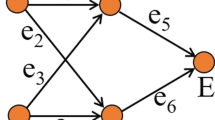
Abstract
Nano scale motility, that is the hallmark of life, emerges when coordinated movements take over from molecular chaos. In biological world, at nanometric size, significant witnesses are present, starting from cellular traffic flows till the organized behaviors of complex molecular machines. Although their origins and evolution are still the objects of investigations, the understanding of physico-chemical mechanisms capable of extracting coordinated movements from the stochastic energy fluctuations of the surrounding liquid is improving. By accounting for the contribution due to the free energy supplied from the hydrolysis of ATP, information theory allows some meaningful in-deepments on processes and mechanisms that contribute to the increase in the complexity present in biological processes. The transfer of some engineering concepts to the biological cells shaped through a set of interacting components with functional, as well as structural relationships, evidences the presence of set-up similar to the ones conventional computers, in which the electronic circuits are able to manage logical expressions. The implications on the emergent synthetic biology are discussed, as well.












(Taken from Benoit Perthame, ENS Paris)

(Taken from: Cohen W, A Computer Scientist’s Guide to Cell Biology: Travelogue from a Stranger in a Strange Land)


(Arranged from Gerd H. G. and Moe Berhens, “Biological Microprocessor, No. 7, April 2013)


Similar content being viewed by others
Abbreviations
- a :
-
Molecular or particle diameter
- \(D = (k_{{\text{B}}} T/3\pi \mu a)\) :
-
Diffusion coefficient
- E :
-
Energy
- Ex:
-
Exergy
- F = U − TS :
-
Helmholtz free energy
- G = H − TS :
-
Gibbs free energy
- H :
-
Enthalpy
- I :
-
Information
- P :
-
Pressure
- Pi:
-
Phosphoric acid
- pi :
-
Probability
- P(x,t):
-
Probability distribution function
- S :
-
Entropy
- t :
-
Time
- T :
-
Temperature
- u :
-
Fluid or particle velocity
- U :
-
Internal energy, characteristic fluid velocity in Eq. (1)
- V :
-
Potential energy
- w(x n/x s):
-
Transition probability between the states n and s
- \(\mu\) :
-
Viscosity
- \(\rho\) :
-
Density
- x,y,z :
-
Cartesian coordinates
- \({\text{d}}r^{3} = {\text{d}}x{\text{d}}y{\text{d}}z\) :
-
Elementary volume in a Cartesian space
- Re = ULρ/μ :
-
Reynolds number
- δ(…):
-
Dirac delta function
- \(\Phi\) :
-
Flux
- \(\nabla\) :
-
Gradient
- \(\nabla^{2} \equiv \nabla \cdot (\nabla )\) :
-
Laplacian
- \(\beta = 1/k_{{\text{B}}} T\) :
-
Being kB the Boltzmann constant
References
Adami C (2012) The use of information theory in evolutionary biology. Ann N Y Acad Sci. 1256(49):65
Adami C, Ofria C, Collier T (1997) Evolution of biological complexity. Proc Nat AcadSci USA 97:4463
Astumian R (2007) Design principles for Brownian molecular machines: how to swim in molasses and walk in a hurricane. PhysChemChemPhys 9:5067
Bray D (2009) Wetware, a computer in every cell. Yale University Press, London
Bushev M (1994) Synergetics, chaos, order and self Organization. World Scientific Publishing, Singapore
Carrà S (2017) Peculiarity and perspectives of catalytic reaction engineering Rend. FisAccLincei. https://doi.org/10.1007/s12210-017-0598-y
Carrà S (2018) Stepping stones to synthetic biology. Springer International Publishing, New York
Carrà S (2020) Reaction kinetics: scientific passion or applicative tool? Rend Fis Acc Lincei. https://doi.org/10.1007/s12210-020-00884-z
Carroll S (2017) The big picture. DUTTON, New York
Cercignani C (1998) Ludwig Boltzmann, the man who trusted atoms. Oxford University Press, Oxford
Chaitin G (2007) Thinking about Godel and Turing: essays on complexity. World Scientific, Singapore
Chaitin G (2012) Proving Darwin making biologic mathematic. Vintage, New York
Chalub F (2002) Kinetics models for Chemotaxis and their Drift. Diffusion limits. https://www.researchgate.net/publication/225454489
Chaudhury S, Cherayil BJ (2006) Complex chemical kinetics in single enzyme molecules. J Chem Phys 125:024904
Danchin A (2009) Bacteria as computers making computers. FEMS Microbiol Rev 33:3–26
Davies P (2019) The demon in the machine. Penguin Random House, London
Einstein A (1905) On the movement of small particles suspended in stationary liquids required by the molecular-kinetic theory of heat. Annalen der Physik (in German) 322(8):549–560
Erbas-Cakmak S (2015) Artificial molecular machines. Chem Rev 2015(115):10081–11020. https://doi.org/10.1021/acs.chemrev.5b00146
Falkowski PG (2015) Life’s engines. Princeton University Press, Princeton
Flack J (2017) Life’s information Hierarchy, in Walker S.I. From matter to Life. Cambridge University Press, Cambridge
Gell-Mann M (1994) The Quark and the Jaguar. Freeman and Company, New York
Gentili PL (2018) The Fuzziness of the molecular world and its perspectives. Molecules 23:2074
Gerd HG et al (2013) The biological microprocessor, or how to build a computer with biological parts. ComputStructBiotechnol J. https://doi.org/10.5936/csbj.201304003
Gerritsma E, Gaspard P (2010) Chemomechanical coupling and stochastic thermodynamics of the F1-ATPase molecular motor with an applied external torque. Biophys Rev Lett 5:163–208
Hanggi P, Talkner P, Borkovec M (1990) Reaction-rate theory: fifty years after Kramers. Rev Modern Phys 62(2):251
Hoffmann P (2012) Life’s Ratchet how molecular machines extract order. Basic Books, New York
Hofstadter R (1979) Gödel, Escher, Bach: an eternal golden braid. Basic Books, New York
Jablonsky J et al (2011) Modeling the Calvin-Benson cycle. BMC Syst Biol 5:18. http://www.biomedcentral.com/1752-0509/5/185.
Lan G, Sun SX (2012) Mechanochemical models of processive molecular motors. MolPhys 110:1017–1034
Landau LD, Lifshitz EM (2009) Fluid mechanics. Butterworth, Oxford
Landauer R (1975) Inadequacy of entropy derivatives in characterizing the steady state. Phys Rev A 18(8):636–638
Lee SY et al (2019) A comprehensive metabolic map for production of bio-based chemicals. Nat Catal 2:18-33. www.nature.com/natcatal
Lovelock J (2019) Novacene. MIT Press, New York
Machta J (1999) Entropy, information, and computation. Am J Phys 67(12):1074–1077
Mayfield J (2013) The engine of complexity, evolution as computation. Columbia University Press, New York
Maynard SJ, Szathmary E (1995) The major transition in evolution. Freeman, Oxford
Mc Clare C (1971) Chemical machines, Maxwell’s demon and living organisms. J TheorBiol 30:1–34
Morton O (2007) Eating the Sun. Fourth Estate, London
Purcell EM (1977) Life at low Reynolds number. Am J Phys 45(1):3–11
Rodnina M, Wintermayer W (2011) The ribosome as a molecular machine: the mechanism of tRNA–mRNA movement in translocation. BiochemSoc Trans 39:658–662. https://doi.org/10.1042/BST0390658
Saunders M, Voth G (2013) Coarse-graining method for computational biology. Annu Rev Biophys 42:73–93
Schuster P (2016) Increase in complexity and information through molecular evolution. Entropy 18:397. https://doi.org/10.3390/e18110397
Sertorio L, Renda E (2009) Ecofisica. BollatiBoringhieri, Torino
Shannon C (1948) A mathematical theory of communication. Bell Syst Tech J 379–423:623–656
Simpson M et al. (2004) Engineering in the biological substrate: information processing in genetic circuits. In: Proceedings of the IEEE, Vol 92, No. 5
Sokolov A et al (2013) Numerical simulation of Chemotaxis models on stationary surfaces. Discrete ContinDynSyst B. https://doi.org/10.3934/dcdsb.2013.18.2689
Spirin A (2002) Ribosome as molecular machine. FEBS Lett 514:2–10
Stephanopoulos G (1998) Metabolic engineering. Academic Press, New York
Stratonovitch R (1992) Nonlinear non-equilibrium thermodynamics, Series in Synergetics. Springer, New York
Vologodskii A (2006) Energy transformation in biological molecular motors. Phys Life Rev 3:119–132
Wall G, Gong M (2001) Onexergy and sustainable development—part 1: conditions and concepts. ExergyInt J 1(3):128–145
Wolpert D (2016) The free energy requirements of biological organisms; implications for evolution. Entropy 18:138. https://doi.org/10.3390/e18040138
Yang H-C, Ge Y-C, Su K-H, Chang C-C, Lin K-C, Aquilanti V, Kasai T (2021) Temperature effect on water dynamics in tetramer phosphofructokinase matrix and the super-Arrhenius respiration rate. Sci Reports 11:1–14
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This paper is the peer-reviewed version of a presentation.at the Conference Statistical thermodynamics and chemical kinetics: far away from equilibrium held at the Accademia Nazionale dei Lincei in Rome, 25–26 June 2019. Program and abstracts at the link Statistical Thermodynamics and Chemical—Manifestazione | Accademia Nazionale dei Lincei.
Rights and permissions
About this article
Cite this article
Carrà, S. At the onset of bio-complexity: microscopic devils, molecular bio-motors, and computing cells. Rend. Fis. Acc. Lincei 32, 215–232 (2021). https://doi.org/10.1007/s12210-020-00971-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12210-020-00971-1