Abstract
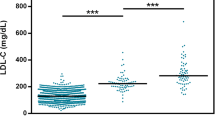
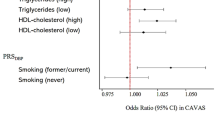
Familial hypercholesterolemia (FH) is an autosomal dominant monogenic disorder characterized by elevated levels of low-density lipoprotein cholesterol (LDL-C) and an increased risk of premature coronary artery disease (CAD). Recently, it has been shown that a high polygenic risk score (PRS) could be an independent risk factor for CAD in FH patients of European ancestry. However, it is uncertain whether PRS is also useful for risk stratification of FH patients in East Asia. We recruited and genotyped clinically diagnosed FH (CDFH) patients from the Kanazawa University Mendelian Disease FH registry and controls from the Shikamachi Health Improvement Practice genome cohort in Japan. We calculated PRS from 3.6 million variants of each participant (imputed from the 1000 Genome phase 3 Asian dataset) for LDL-C (PRSLDLC) using a genome-wide association study summary statistic from the BioBank Japan Project. We assessed the association of PRSLDLC with LDL-C and CAD among and within monogenic FH, mutation negative CDFH, and controls. We tested a total of 1223 participants (376 FH patients, including 173 with monogenic FH and 203 with mutation negative CDFH, and 847 controls) for the analyses. PRSLDLC was significantly higher in mutation negative CDFH patients than in controls (p = 3.1 × 10−13). PRSLDLC was also significantly linked to LDL-C in controls (p trend = 3.6 × 10−4) but not in FH patients. Moreover, we could not detect any association between PRSLDLC and CAD in any of the groups. In conclusion, mutation negative CDFH patients demonstrated significantly higher PRSLDLC than controls. However, PRSLDLC may have little additional effect on LDL-C and CAD among FH patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Harada-Shiba M, Arai H, Oikawa S, Ohta T, Okada T, Okamura T, et al. Guidelines for the management of familial hypercholesterolemia. J Atheroscler Thromb. 2012;19:1043–60.
Kawashiri MA, Hayashi K, Konno T, Fujino N, Ino H, Yamagishi M. Current perspectives in genetic cardiovascular disorders: from basic to clinical aspects. Heart Vessels. 2014;29:129–41.
Tada H, Kawashiri MA, Nohara A, Inazu A, Mabuchi H, Yamagishi M. Impact of clinical signs and genetic diagnosis of familial hypercholesterolaemia on the prevalence of coronary artery disease in patients with severe hypercholesterolaemia. Eur Heart J. 2017;38:1573–9.
Khera AV, Won HH, Peloso GM, Lawson KS, Bartz TM, Deng X, et al. Diagnostic yield and clinical utility of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J Am Coll Cardiol. 2016;67:2578–89.
Mabuchi H. Half a century tales of familial hypercholesterolemia (FH) in Japan. J Atheroscler Thromb. 2017;24:189–207.
Niemi MEK, Martin HC, Rice DL, Gallone G, Gordon S, Kelemen M, et al. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature. 2018;562:268–71.
Choi SW, Mak T, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15:2759–72.
Paquette M, Chong M, Theriault S, Dufour R, Pare G, Baass A. Polygenic risk score predicts prevalence of cardiovascular disease in patients with familial hypercholesterolemia. J Clin Lipido. 2017;11:725–32 e5.
Trinder M, Li X, DeCastro ML, Cermakova L, Sadananda S, Jackson LM, et al. Risk of premature atherosclerotic disease in patients with monogenic versus polygenic familial hypercholesterolemia. J Am Coll Cardiol. 2019;74:512–22.
Sarraju A, Knowles JW. Genetic testing and risk scores: impact on familial hypercholesterolemia. Front Cardiovasc Med. 2019;6:5.
Peloso GM, Nomura A, Khera AV, Chaffin M, Won HH, Ardissino D, et al. Rare protein-truncating variants in APOB, lower low-density lipoprotein cholesterol, and protection against coronary heart disease. Circ Genom Precis Med. 2019;12:e002376.
Nomura A, Emdin CA, Won HH, Peloso GM, Natarajan P, Ardissino D, et al. Heterozygous ABCG5 gene deficiency and risk of coronary artery disease. Circ Genom Precis Med. 2020;13:417–23.
Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018;50:390–400.
Harada-Shiba M, Arai H, Ishigaki Y, Ishibashi S, Okamura T, Ogura M, et al. Guidelines for diagnosis and treatment of familial hypercholesterolemia 2017. J Atheroscler Thromb. 2018;25:751–70.
Nomura A, Won HH, Khera AV, Takeuchi F, Ito K, McCarthy S, et al. Protein-truncating variants at the cholesteryl ester transfer protein gene and risk for coronary heart disease. Circ Res. 2017;121:81–8.
Tada H, Kawashiri MA, Nomura A, Teramoto R, Hosomichi K, Nohara A, et al. Oligogenic familial hypercholesterolemia, LDL cholesterol, and coronary artery disease. J Clin Lipido. 2018;12:1436–44.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics .2010;26:2069–70.
Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–402.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature .2016;536:285–91.
Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607.
Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet. 2014;81:7.23.1–21.
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5.
Mabuchi H, Nohara A, Noguchi T, Kobayashi J, Kawashiri MA, Inoue T, et al. Genotypic and phenotypic features in homozygous familial hypercholesterolemia caused by proprotein convertase subtilisin/kexin type 9 (PCSK9) gain-of-function mutation. Atherosclerosis .2014;236:54–61.
Ohta N, Hori M, Takahashi A, Ogura M, Makino H, Tamanaha T, et al. Proprotein convertase subtilisin/kexin 9 V4I variant with LDLR mutations modifies the phenotype of familial hypercholesterolemia. J Clin Lipido. 2016;10:547–55 e5.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Kawai Y, Mimori T, Kojima K, Nariai N, Danjoh I, Saito R, et al. Japonica array: improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J Hum Genet. 2015;60:581–7.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience .2015;4:7.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9.
Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–97.
Browning BL, Browning SR. Genotype imputation with millions of reference samples. Am J Hum Genet. 2016;98:116–26.
The 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature .2015;526:68–74.
Vilhjalmsson BJ, Yang J, Finucane HK, Gusev A, Lindstrom S, Ripke S, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–92.
Wu H, Forgetta V, Zhou S, Bhatnagar SR, Pare G, Richards JB. A polygenic risk score for low-density lipoprotein cholesterol is associated with risk of ischemic heart disease and enriches for individuals with familial hypercholesterolemia. Circ Genom Precis Med. 2021;14:e003106.
Rieck L, Bardey F, Grenkowitz T, Bertram L, Helmuth J, Mischung C, et al. Mutation spectrum and polygenic score in German patients with familial hypercholesterolemia. Clin Genet. 2020;98:457–67.
Trinder M, Paquette M, Cermakova L, Ban MR, Hegele RA, Baass A, et al. Polygenic contribution to low-density lipoprotein cholesterol levels and cardiovascular risk in monogenic familial hypercholesterolemia. Circ Genom Precis Med. 2020;13:515–23.
Trinder M, Francis GA, Brunham LR. Association of monogenic vs polygenic hypercholesterolemia with risk of atherosclerotic cardiovascular disease. JAMA Cardiol. 2020;5:390–99.
Ripatti P, Ramo JT, Mars NJ, Fu Y, Lin J, Soderlund S, et al. Polygenic hyperlipidemias and coronary artery disease risk. Circ Genom Precis Med. 2020;13:e002725.
Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM, et al. Predictive accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA .2020;323:636–45.
Mosley JD, Gupta DK, Tan J, Yao J, Wells QS, Shaffer CM, et al. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA. 2020;323:627–35.
Acknowledgements
We thank all the participants and medical staff that were involved in this study. Also, this study was supported in part by the Japan Heart Foundation Research Grant, Astellas Foundation for Research on Metabolic Disorders, Japan Research Foundation for Clinical Pharmacology, Japan Cardiovascular Research Foundation (The Bayer Scholarship for Cardiovascular Research), The Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the KAKEN Grant-in-Aid for Scientific Research (C) (19K08553), (B) (21H03179), and for Challenging Research (Exploratory) (19K22753).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
AN received consulting fees from CureApp, Inc. and was a co-founder of the CureApp Institute. The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Nomura, A., Sato, T., Tada, H. et al. Polygenic risk scores for low-density lipoprotein cholesterol and familial hypercholesterolemia. J Hum Genet 66, 1079–1087 (2021). https://doi.org/10.1038/s10038-021-00929-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-021-00929-7
This article is cited by
-
Genetic factors associated with serum amylase in a Japanese population: combined analysis of copy-number and single-nucleotide variants
Journal of Human Genetics (2023)
-
Applicability of polygenic risk scores in endometriosis clinical presentation
BMC Women's Health (2022)
-
Common genetic variation associated with Mendelian disease severity revealed through cryptic phenotype analysis
Nature Communications (2022)