Abstract
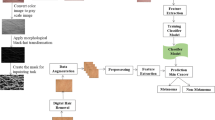
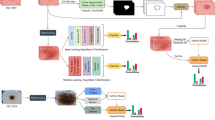
ResNet50 and VGG-16 models are introduced in this paper with different strategies, with and without preprocessing and with and without Support Vector Machine (SVM). Moreover, both transfer learning and data augmentation are used to solve the problem of lack of tagged data. The fully connected (FC) layer is replaced by the SVM classifier leading to better accuracy. In addition, in our work, we utilize the median filter, contrast enhancement and edge detection, which based on four main steps: noise removal, gradient smoothed image calculations, non-highest suppression and hysteresis thresholding. Also, the k-fold cross validation is performed to authenticate our model’s performance. Three data sets: ISIC 2017 MNIST-HAM10000 and ISBI 2016 are utilized in our proposed work. It is observed that the proposed technique of employing ResNet50 hybridized with SVM achieves the best performance, specifically with the ISIC2017 dataset, producing 99.19% accuracy, 99.32% area under the curve (AUC), 98.98% sensitivity, 98.78% precision, 98.88% F1 score and 2.6988 s computational time.








Similar content being viewed by others
References
Abbas Q, Sadaf M, Akram A (2016) Prediction of dermoscopy patterns for recognition of both melanocytic and non-melanocytic skin lesions. Computers 5(13):1–16
Albahar MA (2019) Skin lesion classification using convolutional neural network with novel regularizer. IEEE Access 7:38306–38313
Brinker TJ, Hekler A, Utikal JS, Grabe N, Schadendorf D, Klode J, Berking C, Steeb T, Alexander HE, Kalle CV (2018) Skin cancer classification using convolutional neural networks: systematic review. J Med Internet Res 20(10):e11936
Canny J (1986) A computational approach to edge detection. IEEE Trans Pattern Anal Mach Intell PAMI-8(6):679–698
Cervantes J, Garcia-Lamont F, Rodriguez-Mazahua L, Lopez A (2020) A comprehensive survey on support vector machine classification: applications, challenges and trends. Neurocomputing 408:189–215
Dalila F, Zohra A, Reda K, Hocine C (2017) Segmentation and classification of melanoma and benign skin lesions. Optik 140:749–761
Demyanov S, Chakravorty R, Abedini M, Halpern A, Garnavi R (2016) Classification of dermoscopy patterns using deep convolutional neural networks. In: 2016 IEEE 13th international symposium on biomedical imaging (ISBI). Czech Republic, Prague, pp 364–368
Eltrass A, Salama M (2020) Fully automated scheme for computer-aided detection and breast cancer diagnosis using digitised mammograms. IET Image Process 14(3):495–505
Feng X, Hongxun Y, Shengping Z (2019) An efficient way to refine DenseNet. SIViP 13(5):959–965
Firoz R, Ali M, Khan M, Hossain M, Islam M, Shahinuzzaman M (2016) Medical imageenhancement using morphological transformation. J Data Anal Inf Process 4:1–12.https://doi.org/10.4236/jdaip.2016.41001
Hardie R, Ali R, Silva MSD, Kebede TM (2018) Skin lesion segmentation and classification for ISIC 2018 using traditional classifiers with handcrafted features. arXiv:1807.07001 [eess.IV]
Huang T, Yang G, Tang G (1979) A fast two-dimensional median filtering algorithm. IEEE Trans Acoust Speech Signal Process 27(1):13–18
ISIS (n.d.) Archive [electronic resource], Kitware, Inc., https://isic-archive.com/(accessed January 24, 2020).
K. Inc. (2019) Skin Cancer MNIST: HAM10000. Available: https://www.kaggle.com/kmader/skin-cancer-mnist-ham10000/version/2 (accessed January 24, 2020).
Kasmi R, Mokrani K (2016) Classification of malignant and benign skin lesions: implementation of automatic ABCD rule. IET Image Process 10(6):448–455
Khan MA, Akram T, Sharif M, Shahzad A, Aurangzeb K, Alhussein M, Haider SI, Altamrah A (2019) An implementation of normal distribution based segmentation and entropy controlled features selection for skin lesion detection and classification. BMC Cancer, 18(1), 638, 2018.Kingdom, pp. 1229–1233. https://doi.org/10.1186/s12885-018-4465-8
Khan MA, Sharif M, Akram T, Bukhari SAC, Nayak RS (2020) Developed Newton-Raphson based deep features selection framework for skin lesion recognition. Pattern Recogn Lett 129:293–303
Mahbod A, Schaefer G, Wang C, Ecker R, Ellinge I (2019) Skin lesion classification using hybrid deep neural networks. In ICASSP 2019 IEEE international conference on acoustics, speech and signal processing (ICASSP’2019), Brighton, UK. https://doi.org/10.1109/ICASSP.2019.8683352
Majtner T, Yildirim-Yayilgan S, Hardeberg JY (2016) Combining deep learning and hand-crafted features for skin lesion classification. In 2016 IEEE sixth international conference on image processing theory, tools and applications (IPTA), Oulu, Finland, pp. 1–6. https://doi.org/10.1109/IPTA.2016.7821017
Matsunaga K, Hamada A, Minagawa A, Koga H (2017) Image classification of melanoma, nevus and seborrheic keratosis by deep neural network ensemble. arXiv preprint arXiv:1703.03108
Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Pham BT, Jaafari A, Prakash I, Bui DT (2019) A novel hybrid intelligent model of support vector machines and the multi-boost ensemble for landslide susceptibility modeling. Bull Eng Geol Environ 78(4):2865–2886
Rembielak A, Ajithkumar T (2019) Non-melanoma skin cancer–an underestimated global health threat. Clin Oncol 31(11):735–737
Salama MS, Eltrass AS, Elkamchouchi HM (2018) An improved approach for computer-aided diagnosis of breast cancer in digital mammography. In IEEE International Symposium on Medical Measurements and Applications, Rome, Italy, 1–5
Salamon J, Bello JP (2017) Deep convolutional neural networks and data augmentation for environmental sound classification. IEEE Signal Process Lett 24(3):279–283
Santos MO (2018) Estimate: cancer incidence in Brazil. Revista Brasileira de Cancerologia 64(1):119–120
Serte S, Demirel H (2019) Gabor wavelet-based deep learning for skin lesion classification. Comput Biol Med 113:103423
Shin HC, Roth HR, Mingchen G, Lu L, Ziyue X, Nogues I, Jianhua Y, Mollura D, Summers RM (2016) Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35(5):1285–1298
Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. J Big Data 6(1):60
Sofaer HR, Hoeting JA, Jarnevich CS (2019) The area under the precision-recall curve as a performance metric for rare binary events. Methods Ecol Evol 10:565–577
Srinivasan K, Cherukuri AK, Vincent DR, Garg A, Chen B-Y (2019) An efficient implementation of artificial neural networks with K-fold cross-validation for process optimization, J. Internet Technol 20:1213–1225
Tschandl P, Codella N, Akay NB, Argenziano G, Braun PR, Cabo H, Gutman D, Halpern A, Helba B, Wellenhof RH, Lallas A (2019) Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Lancet Oncol 20(7):938–947
Vasconcelos CN, Vasconcelos BN (2017) Experiments using deep learning for dermoscopy image analysis. Pattern Recogn Lett 139(2020):95–103
Yilmaz E, Trocan M (2020) Benign and malignant skin lesion classification comparison for three deep-learning architectures. In: Nguyen N, Jearanaitanakij K, Selamat A, Trawiński B, Chittayasothorn S (eds) Intelligent information and database systems. ACIIDS. Lecture notes in computer science, 12033. Springer, Cham
Yoshida T, Celebi ME, Schaefer G, Iyatomi H (2016) Simple and effective pre-processing for automated melanoma discrimination based on cytological findings. In 2016 IEEE international conference on big data (big data), Washington, USA, pp. 3439–3442. https://doi.org/10.1109/BigData.2016.7841005
Yu L, Chen H, Dou Q, Qin J, Heng PA (2016) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging 36(4):994–1004
Yu D, Wang H, Chen P, Wei Z (2014) Mixed pooling for convolutional neural networks. In: Miao D, Pedrycz W, Ślȩzak D, Peters G, Hu Q,Wang R (eds) Rough Sets and Knowledge Technology. RSKT 2014. Lecture Notes in Computer Science, vol. 8818. Springer, Cham. https://doi.org/10.1007/978-3-319-11740-9_34
Yüksel M, Emin, Borlu M (2009) Accurate segmentation of dermoscopic images by image thresholding based on type-2 fuzzy logic. IEEE Trans Fuzzy Syst 17(4):976–982
Zachary HR, Secrest AM (2019) Public health implications of google searches for sunscreen, sunburn, skin cancer, and melanoma in the United States. Am J Health Promot 33(4):611–615
Zhang J, Xie Y, Xia Y, Shen C (2019) Attention residual learning for skin lesion classification. IEEE Trans Med Imaging 38(9):2092–2103
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Wessam M. Salama and Moustafa H. Aly declare that there is no conflict of interest.
Human and animal studies
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
The article has no part or work that needs informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Salamaa, W.M., Aly, M.H. Deep learning design for benign and malignant classification of skin lesions: a new approach. Multimed Tools Appl 80, 26795–26811 (2021). https://doi.org/10.1007/s11042-021-11000-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-11000-0