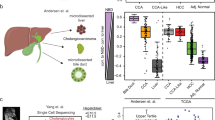
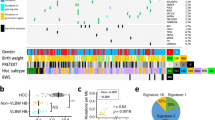
Abstract
Hepatocellular carcinoma (HCC) is a global health issue and the fourth leading cause of cancer deaths worldwide. Large-scale HCC genome sequencing analyses have identified core drivers (TERT, TP53, and CTNNB1/AXIN1) as initial molecular events, and other low-frequent drivers that include therapeutically targetable ones. The recent genetic analysis uncovered a distinctive driver gene landscape in precancerous lesions, arguing a discontinuous process at early HCC development. In advanced tumors, intra-tumoral heterogeneity through clonal evolution processes is common, and it displays clear geographic segregation genetically and epigenetically. Diverse epidemiological risk factors for HCC mirrors heterogeneous mutational processes among patient cohorts with distinctive ethnicity, environmental exposures, and lifestyles. The genetic information of individual tumors has been utilized for optimizing treatments, early diagnosis, and monitoring recurrence. It will expand the opportunity for screening high-risk populations, thereby preventing hepatocarcinogenesis in the near future.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–17.
Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340–9.
Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–73.
Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–11.
Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500–9.
Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–41.
Nault JC, Mallet M, Pilati C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218.
Midorikawa Y, Yamamoto S, Tatsuno K, et al. Accumulation of molecular aberrations distinctive to hepatocellular carcinoma progression. Cancer Res. 2020;80:3810–9.
St Pierre R, Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev. 2017;42:56–67.
Sun X, Wang SC, Wei Y, et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell. 2017;32:574–89.
Ogiwara H, Takahashi K, Sasaki M, et al. Targeting the vulnerability of glutathione metabolism in ARID1A-deficient cancers. Cancer Cell. 2019;35:177–90.
Sankar TS, Wastuwidyaningtyas BD, Dong Y, et al. The nature of mutations induced by replication–transcription collisions. Nature. 2016;535:178–81.
Zhu M, Lu T, Jia Y, et al. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell. 2019;177:608–21.
Kim SK, Takeda H, Takai A, et al. Comprehensive analysis of genetic aberrations linked to tumorigenesis in regenerative nodules of liver cirrhosis. J Gastroenterol. 2019;54:628–40.
Brunner SF, Roberts ND, Wylie LA, et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574:538–42.
Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101.
Ng AWT, Poon SL, Huang MN, et al. Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci Transl Med. 2017;9:eaan6446.
Lu ZN, Luo Q, Zhao LN, et al. The mutational features of aristolochic acid-induced mouse and human liver cancers. Hepatology. 2020;71:929–42.
Hama N, Totoki Y, Miura F, et al. Epigenetic landscape influences the liver cancer genome architecture. Nat Commun. 2018;9:1643.
Sung WK, Zheng H, Li S, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–9.
Zhao LH, Liu X, Yan HX, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7:12992.
Furuta M, Tanaka H, Shiraishi Y, et al. Characterization of HBV integration patterns and timing in liver cancer and HBV-infected livers. Oncotarget. 2018;9:25075–88.
Nault JC, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47:1187–93.
Park KJ, Lee J, Park JH, et al. Adeno-associated virus 2-mediated hepatocellular carcinoma is very rare in Korean patients. Ann Lab Med. 2016;36:469–74.
Tatsuno K, Midorikawa Y, Takayama T, et al. Impact of AAV2 and hepatitis B virus integration into genome on development of hepatocellular carcinoma in patients with prior hepatitis B virus infection. Clin Cancer Res. 2019;25:6217–27.
La Bella T, Imbeaud S, Peneau C, et al. Adeno-associated virus in the liver: natural history and consequences in tumour development. Gut. 2020;69:737–47.
Kudo M. Early hepatocellular carcinoma: definition and diagnosis. Liver Cancer. 2013;2:69–72.
Takeda H, Takai A, Kumagai K, et al. Multiregional whole-genome sequencing of hepatocellular carcinoma with nodule-in-nodule appearance reveals stepwise cancer evolution. J Pathol. 2020;252:398–410.
Zhai W, Lim TK, Zhang T, et al. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat Commun. 2017;8:4565.
Ding X, He M, Chan AWH, et al. Genomic and epigenomic features of primary and recurrent hepatocellular carcinomas. Gastroenterology. 2019;157:1630–45.e6.
Furuta M, Ueno M, Fujimoto A, et al. Whole genome sequencing discriminates hepatocellular carcinoma with intrahepatic metastasis from multi-centric tumors. J Hepatol. 2017;66:363–73.
Cai Z, Chen G, Zeng Y, et al. Comprehensive liquid profiling of circulating tumor DNA and protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin Cancer Res. 2019;25:5284–94.
Qu C, Wang Y, Wang P, et al. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAg-seropositive individuals by liquid biopsy. Proc Natl Acad Sci USA. 2019;116:6308–12.
Hadi K, Yao X, Behr JM, et al. Distinct classes of complex structural variation uncovered across thousands of cancer genome graphs. Cell. 2020;183:197–210.
Shibata T, Arai Y, Totoki Y. Molecular genomic landscapes of hepatobiliary cancer. Cancer Sci. 2018;109:1282–91.
Acknowledgements
This work was supported in part by the Practical Research for Innovative Cancer Control from the Japan Agency for Medical Research and Development (AMED, JP20ck0106547h0001), National Cancer Center Research and Development Funds (2020-A-7).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shibata, T. Genomic landscape of hepatocarcinogenesis. J Hum Genet 66, 845–851 (2021). https://doi.org/10.1038/s10038-021-00928-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-021-00928-8
This article is cited by
-
Current status and issues in genomic analysis using EUS-FNA/FNB specimens in hepatobiliary–pancreatic cancers
Journal of Gastroenterology (2023)