Abstract
Both genetic drift and divergent selection are predicted to be drivers of population differentiation across patchy habitats, but the extent to which these forces act on natural populations to shape traits is strongly affected by species’ ecological features. In this study, we infer the genomic structure of Pitcairnia lanuginosa, a widespread herbaceous perennial plant with a patchy distribution. We sampled populations in the Brazilian Cerrado and the Central Andean Yungas and discovered and genotyped SNP markers using double-digest restriction-site associated DNA sequencing. In addition, we analyzed ecophysiological traits obtained from a common garden experiment and compared patterns of phenotypic and genetic divergence (PST–FST comparisons) in a subset of populations from the Cerrado. Our results from molecular analyses pointed to extremely low genetic diversity and a remarkable population differentiation, supporting a major role of genetic drift. Approximately 0.3% of genotyped SNPs were flagged as differentiation outliers by at least two distinct methods, and Bayesian generalized linear mixed models revealed a signature of isolation by environment in addition to isolation by distance for high-differentiation outlier SNPs among the Cerrado populations. PST–FST comparisons suggested divergent selection on two ecophysiological traits linked to drought tolerance. We showed that these traits vary among populations, although without any particular macro-spatial pattern, suggesting local adaptation to differences in micro-habitats. Our study shows that selection might be a relevant force, particularly for traits involved in drought stress, even for populations experiencing strong drift, which improves our knowledge on eco-evolutionary processes acting on non-continuously distributed species.
Similar content being viewed by others
Introduction
The comparison of neutral and adaptive genomic variation has allowed valuable insights into the evolutionary processes driving genetic structure in natural populations (Orsini et al. 2013; Andrews et al. 2016). Species with naturally isolated populations represent particularly interesting systems to infer the relative roles of genetic drift and natural selection for population differentiation, an important process in biological diversification. Genetic studies on such non-continuously distributed systems have shown that restricted gene flow among populations often results in strong genetic drift (e.g. Ortego et al. 2010; Palma-Silva et al. 2011; Nistelberger et al. 2015), but the importance of selection for genetic differentiation and local adaptation of these species has only recently been tested using molecular markers (e.g., Funk et al. 2016; Perrier et al. 2017; Wang et al. 2017; Prentice et al. 2017). Although both evolutionary forces are predicted to be important and strongly affected by the effective population size and population persistence through time, selection differentially affects traits and their underlying genetic variation (Flood and Hancock 2017). The smaller the effective population size and migration rates among populations, the larger the effects of stochastic processes and the lower the effectiveness of selection (Lande and Barrowclough 1987; Charlesworth 2009). Furthermore, local colonization-extinction dynamics may enhance the effects of drift across isolated populations. Although we lack empirical evidence on genetic drift acting as a sole driver of speciation processes, e.g., through relaxing purifying selection thus allowing genetic incompatibilities to accumulate between isolated populations (Coyne and Orr 2004; Sobel et al. 2010), the role of selection is often hard to demonstrate when drift is strong. The identification of the mechanisms responsible for population differentiation across patchy habitats may, therefore, shed light on the potential but still controversial role of drift and on the relative importance of selection on speciation processes.
The ability of species to disperse and establish in habitats under distinct climate conditions is directly linked to the interplay between drift and selection. While many species show evidence of niche differentiation, others have strict ecological requirements and establish in regions with similar environmental conditions (i.e., niche conservatism) (McCormack et al., 2010; Bacon et al. 2017; Baranzelli et al. 2018). The ability of organisms to adapt to new environmental conditions is determined by de novo mutations or by standing genetic variation, but adaptation from standing variation is likely to be faster (Barrett and Schluter 2008). Depending on the magnitude of selection and the effective population size, drift may overwhelm divergent selection on new beneficial mutations underlying a particular trait in small populations colonizing new habitats (Wright 1951; Leimu and Fischer 2008). Although population size greatly influences persistence, small founder populations may still hold considerable standing genetic variation on which selection could act (Agashe et al. 2011). Understanding the evolutionary strategy that allows, or not, non-continuously distributed species potentially subject to strong founder events to increase their ecological range toward distinct ecological conditions requires coupling genetic and ecological information.
For the highly heterogeneous Neotropics, an increasing number of genetic studies in widespread species has improved our understanding of diversification across biomes (Antonelli et al. 2018), but whether differentiation within species is accompanied or not by local adaptation has been less explored (but see Bacon et al. 2017). The application of high throughput sequencing to Neotropical systems promises to allow novel insights into diversification events and the history of biomes (e.g., Nadeau et al. 2013; Harvey and Brumfield 2015; Ebel et al. 2015). Indeed, the increasing number of loci generated by these sequencing technologies and the constant development of computational methods have allowed for more powerful inferences regarding genetic structure in non-model species (Davey et al. 2011; Narum et al. 2013), and have also greatly improved our ability to distinguish neutral from adaptive processes (Andrews et al. 2016). Approaches for identifying locally adaptive candidate loci rely on the identification of markers with unusually high genetic differentiation among populations or searching for correlations between allele frequencies and environmental information (Hoban et al. 2016). Moreover, when phenotypic information is available at the population level, phenotypic divergence (Pst) can be compared to neutral genetic divergence (Fst) to give insights into the degree of differentiation caused by selective versus neutral processes (Pst–Fst comparisons; Brommer 2011; Leinonen et al. 2013), particularly when controlling for environmental variation is feasible. Nevertheless, there are still challenges associated with identifying sufficient genetic variation to detect loci under selection and to perform genotype-environment or genotype-phenotype associations (Leinonen et al. 2013; Lowry et al. 2017), particularly in species with restricted gene flow (Perrier et al. 2017).
In this study, we quantify natural selection on selected ecophysiological traits and infer the relative roles of genetic drift and natural selection on population differentiation in Pitcairnia lanuginosa Ruiz and Pav. This species has a widespread distribution in tropical regions of South America, with small populations typically scattered across space and associated with riparian forests in the Brazilian Cerrado and the Central Andean Yungas (Fig. 1). A phylogeographic study employing microsatellite markers and Sanger sequencing has suggested that the divergence between lineages occupying the Cerrado and Yungas is likely caused by past dispersal rather than vicariance (Leal et al. 2019). The authors found low levels of genetic diversity (mean HE = 0.205) and strong population structure (global FST = 0.73), likely resulting from limited gene flow due to the lack of dispersal traits in seeds, small populations, and extremely high rates of spontaneous self-fertilization in this species. Despite the strong effect of drift suggested by these findings, adaptive processes may also be important to explain divergence patterns of P. lanuginosa considering that populations are potentially submitted to contrasting macroclimatic conditions across the large-scale latitudinal and longitudinal distribution range of the species. Indeed, P. lanuginosa (here considered synonymous of P. burchellii Mez; Smith and Downs 1974) exhibits physiological and anatomical characteristics to cope with severe drought stress (Vieira et al. 2017a, b), which may be important for local adaptation to the greater seasonality in the Cerrado, a savannah biome with wet summers and dry winters, when compared to the Central Andean Yungas, a mesic forest biome. Nevertheless, the extent to which this strong genetic structure could be interpreted in terms of adaptive processes can be strongly affected by the species mating system (Glémin 2007; Wright et al. 2013). The increased self-fertilization in P. lanuginosa, for instance, may reduce the effective population sizes of populations, thereby enhancing genetic drift. Under strong drift, adaptive alleles can also be lost, limiting the process of local adaptation (Charlesworth and Charlesworth 1987; Wright et al. 2013).
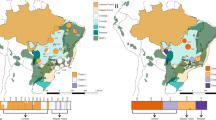
A Geographic distribution of sampled populations assigned to genetic clusters as detected by INSTRUCT. Black dots represent species-occurrences retrieved from Global Biodiversity Information Facility (GBIF) and SpeciesLink. B Bar plot depicting the INSTRUCT clustering for 19 populations of the species. Selfing rates per cluster are shown above the barplot. Each vertical bar corresponds to one individual and colored segments to the proportion of assignment to clusters, each color corresponding to a distinct cluster.
Here we take advantage of hundreds of SNP markers derived from double-digest restriction-site associated DNA sequencing (ddRAD-seq, Peterson et al. 2012), and information on ecophysiological traits associated with environmental variation, to investigate the relative roles of neutral and adaptive processes in causing the strong genetic differentiation in P. lanuginosa. Specifically, we aim to: (1) investigate whether the presumed low effective population sizes resulting from isolation and high levels of inbreeding could preclude local adaptation for ecophysiological traits linked to drought, heat, and light stress in this species; (2) assess the relative importance of Isolation by Distance (IBD) and Isolation by Environment (IBE) on the patterns of genetic structure in this species. The wide yet patchy distribution in the Neotropics makes P. lanuginosa a particularly interesting model to investigate how ecological processes affect local adaptation. The information on the genomic and trait variation in this species may thereby shed light on eco-evolutionary processes acting on non-continuously distributed species.
Material and methods
Model species and population sampling
P. lanuginosa is an iteroparous perennial forb self-compatible and capable of intrafloral autonomous self-pollination (Leal 2018). Populations of this species are generally small and spatially restricted, composed of one to a few dozen individuals that are mostly found near streams and waterfalls (personal observation). We sampled 19 populations of P. lanuginosa across most of its geographic range (Table 1, Fig. 1). The sampling covers the Brazilian Cerrado (savanna biome, including transition zones with the Atlantic Forest and Amazonia) and the Central Andean Yungas (Amazonia-Andes transition) (Fig. 1). Adult specimens from a subset of eight Cerrado populations were transported from the field to the experimental greenhouse of São Paulo State University (UNESP), Rio Claro, Brazil (see Table 1), and planted into pots to grow under controlled conditions. Herbarium specimens were deposited within the herbarium HBRC, UNESP, Rio Claro, Brazil.
Ecophysiological traits
To detect differences in plant responses to light, heat and water stresses among populations, we conducted ecophysiological trait measurements in a total of 24 individuals (herein genets) from eight out of the 19 P. lanuginosa populations sampled (three genets/population); these eight populations represent the extent of the geographic distribution of the species in the Cerrado (Fig. 1; Table 1). Adult plants were sampled in the field and acclimated in the greenhouse for at least 1 year (from May 2014 to May 2015). We separated each genet into two or more ramets to grow in distinct pots with the same substrate composition for an additional acclimation period of 3 months prior to the experiments. We then selected 48 ramets of similar size lacking inflorescences and submitted them to the following treatments (24 ramets/treatment): (A) control, consisting in watering pots every 2 days (about 80 mL of water, which corresponded to field capacity); and (B) water shortage, consisting in no watering for 30 days. We measured ecophysiological traits based on photosynthetic responses of leaves in August 2016 (during the dry season).
We selected an area of ca. 2 cm2 on a fully developed leaf on each ramet from the control treatment to measure the photosynthetic response to light (i.e. rapid light curve) and to heat stress using a modulate fluorometer (MINI-PAM-II, Walz). We calculated the following parameters: upper critical thermal limit (CTmax), sensitivity to temperature change (z), maximum relative electron transport rate (ETRmax), initial slope of the curve of light-response (α), light saturation coefficient (Ik), and maximal photochemical efficiency (Fv/Fm). To calculate heat tolerance parameters CTmax and z, we followed the framework described by Rezende et al. (2014), which considers the intensity and duration of thermal stress to drive the individual ‘thermal tolerance landscape’, or individual capacity to withstand heat at any given temperature. We followed procedures described by Godoy et al. (2011), and modified by Chaves et al. (2018), to measure the critical time duration under a static stressful temperature that promotes a 50% decay of the initial Fv/Fm (D50) in each leaf sample. To draw the ‘thermal tolerance landscape’ of each ramet, we calculated D50 under four distinct temperatures (40, 45, 50, and 60 °C) and performed a linear regression to measure CTmax and z parameters according to Rezende et al. (2014). Measures of Fv/Fm at each temperature were simultaneously carried out using a thermal-gradient block designed by Labouriau (1977) and adapted by Cardoso (2010). Finally, we used the decay of each heat and light parameter (i.e., values for ramets under controlled conditions minus the values under water shortage, divided by values under controlled conditions) as proxies of drought adaptation. Values close to one indicate little difference between treatments, i.e., tolerance to drought stress.
DNA extraction and ddRAD sequencing
Total genomic DNA was extracted from leaf samples of 115 individuals from 19 populations (Table 1) using the Qiagen DNA Plant Mini kit (Qiagen, Finland). Sample quality and concentration were checked on 1% agarose gels and using a NanoDrop 2000 spectrophotometer (Thermo Scientific) and normalized to 60 ng/μl as quantified by the Qubit dsDNA BR assay (Invitrogen). The ddRAD libraries were prepared and sequenced on the Ion Torrent Proton platform using a modified version of Peterson et al. 2012 protocol at the Genome Transcriptome Facility of Bordeaux (INRA, Cestas, France). In brief, we digested 50 μl (300 ng total) of genomic DNA using rare- and frequent-cutter restriction enzymes (PstI and MspI, New England Biosciences) at 37 °C for 2 h and inactivated them at 80 °C for 20 min. After bead-purification, a P1 adaptor (the same for all samples) and a unique barcode adaptor were added to each sample, and ligation was performed at 22 °C for 2 h followed by inactivation at 65 °C for 20 min. We then used a qPCR to quantify each sample before normalizing and pooling samples in a 48-plex. We employed an automated size-selection technology (Pippin, Sage Science) to select fragments with the expected size (270–290 pb). Each library was then amplified by PCR, quantified at the Agilent 2100 Bioanalyzer, and sequenced on the Ion Torrent Proton P1v2 chip.
De novo assembly
We analyzed raw read data using PYRAD v3.0.5 software (Eaton 2014). This software assembles short read RAD-seq (or similar) data using a clustering algorithm that allows for indel variation within and between samples and incomplete overlap among reads (Eaton 2014). This particularity makes the method suitable for highly structured populations. Following preliminary testing, we settled the parameters as follows: a maximum number of sites with quality score <20 (NQual): 5, clustering threshold (Wclust): 0.85, minimum coverage for a cluster (Mindepth): 6, and maximum individuals with a shared heterozygous site (MaxSH): 3. All other parameters were kept at default values. PYRAD was run on the GenoToul bioinformatics facility (INRA, Toulouse, France). We then employed VCFTOOLS v0.1.13 (Danecek et al. 2011) to retain loci with a minimum of 30% of individuals sequenced and removed any individual with genotypes for fewer than 30% of loci. These parameters were chosen to recover a maximum number of loci based on the biology of P. lanuginosa, a frequently selfing species in highly structured populations (Leal et al. 2019). Although our missing data criteria fall below what is typically used for RAD-seq data, studies have shown that even lower missing data cutoffs lead to correct inferences (Chattopadhyay et al. 2014; Huang and Knowles 2016; Hodel et al. 2017). Because of the known long-term divergence between Cerrado and Central Yungas populations (ca. 3 Mya; Leal et al. 2019), another data matrix including only samples from the Cerrado was separately build using PYRAD and then filtered by VCFTOOLS according to the same criteria. VCF files were converted to various formats using PGDSPIDER v2.0.7.2 (Lischer and Excoffier 2012) to perform further analyses.
Genomic diversity and differentiation
We calculated diversity indices, such as the proportion of polymorphic sites (PL), private alleles (PA), observed (HO) and expected (HE) heterozygosities and nucleotide diversity for variant sites (π) using the ‘populations’ module of STACKS v1.35 (Catchen et al. 2013).
We estimated pairwise FST (Weir and Cockerham 1984) and G′ST (Hedrick 2007) among sampled populations using the R package ‘diveRsity’ (Keenan et al. 2013). A neighbor-net unrooted phylogenetic network was inferred with the software SPLITSTREE4 using uncorrected p-distances and a bootstrap procedure to assess confidence (Huson and Bryant 2006). We further assessed patterns of genomic structure employing the Bayesian approach implemented in INSTRUCT (Gao et al. 2007). This software clusters individuals into subpopulations and allows estimating the self-fertilization rates or inbreeding coefficients both at the population level and at the individual level (Gao et al. 2007), and is thus especially suited for species that reproduce through self-fertilization. INSTRUCT was run for K = 1 to K = 20, for five independent chains, each with 200,000 iterations and 50,000 burn-in steps, with a thinning interval of ten steps, using the joint inference of population selfing rate and population sub-structure mode. We chose the best K based on the deviance information criterion (DIC) and used OBSTRUCT (Gayevskiy et al. 2014) to plot averaged results among chains. We then used ARLEQUIN 3.5 (Excoffier and Lischer 2010) to test the partition of genetic diversity within and between congruent groups revealed by STRUCTURE and INSTRUCT using an analysis of molecular variance (AMOVA).
Detection of outlier loci
We employed three different approaches to identify candidate SNPs bearing a signature of selection. The aim was to identify such loci without considering environmental variables (see next section), i.e., based solely on their population genetic signature of selection. This analysis was performed across the entire distribution of P. lanuginosa and among populations sampled in the Cerrado, using the independently assembled data sets. First, we detected outliers based on locus-specific FST using the program BAYESCAN v2.1 (Foll and Gaggiotti 2008) with prior odds of 10 and a false discovery rate (FDR) of 0.05. After 20 pilot runs of 5000 iterations, we ran a total of 100,000 MCMC iterations with a burn-in of 50,000 iterations and a thinning interval of 10. BAYESCAN examines pairwise FST between populations for each locus and decomposes the coefficients into a population-specific component (β), and a locus-specific component (α) using logistic regression (Foll and Gaggiotti 2008). The detected outlier loci with positive α values are suggested to undergo diversifying selection, while those with a negative α are suggested to be under balancing or purifying selection. Diagnostics of the log likelihoods and FST values were checked using the ‘CODA’ R-package (Plummer et al. 2006). To complement this analysis, we used two other differentiation-based methods to identify outlier SNPs that account for population structure: a principal components‐based method implemented in the R-package ‘pcadapt’ (Luu et al. 2017) and a Bayesian method implemented in BAYPASS (Gautier 2015). For the PCADAPT analysis, we assessed the optimal number of principal components (K) from 1 to 20 and inspected the resulting scree plots to retain K = 5, K = 4, and K = 2 for the data sets consisting of all sampled populations and a subset of populations sampled either in the Cerrado or in the Andean Yungas, respectively. For each SNP, deviation from the mean population structure was estimated based on the Mahalanobis distance D that summarizes regression of the SNP by the K principal components. D values were converted to p values and we used an FDR of 0.05 to obtain a list of outlier SNPs. For BAYPASS analysis, we calculated the correlation matrix Ω and estimate XTX, an FST-like statistic, for each individual SNP under the core model using default settings. We simulated pseudo‐observed data sets (PODs) to calibrate the XTX values and used a 95% threshold, as determined from PODs, to discriminate between outlier candidates and neutral SNPs.
Environment–genotype associations
We first tested for the correlation among predictors, i.e. geographical and environmental distances based on climate, soil, and elevation, for populations within the Cerrado with Mantel tests implemented in the R package ‘adegenet’ (Jombart 2008) with 1000 permutations. The geographic matrix consisted of the natural logarithm of Euclidian distances among populations (hereafter, GEO matrix), while the elevation matrix consisted of pairwise differences between the registered elevation values for sampled populations (ELEV matrix). The environmental distances were derived from Euclidean distances among populations along the first and second axis of a principal component analysis (PCA) summarizing (1) 19 WorldClim variables (Hijmans et al. 2005) at a resolution of 30 arc seconds (CLIM matrix) and (2) 10 chemical and physical soil properties downloaded from SoilGrids (Hengl et al. 2017) at a resolution of 250 m and a depth of 0–5 cm (Hengl et al. 2017) (SOIL matrix). The first and second axes of these PCAs for climate and soil data accounted for 70.90% and 62.29% of the data variances, respectively. For the climate data, the PCA axes were mostly correlated with the temperature of the coldest quarter and with the precipitation of the wettest quarter (Table S1). For the soil data, axes 1 and 2 were mostly correlated with the bulk density and proportion of clay particles (Table S2).
We used a Bayesian generalized linear mixed modeling (GLMM) approach to test whether genetic differentiation among the Cerrado populations at the neutral genomic fraction and at each outlier SNP is driven by geographic distance or environmental differences, as determined by variation in temperature and precipitation, elevation or soil. This approach accounts for the non independence of values resulting from the use of pairwise matrices of population‐level metrics and can distinguish the effects of multiple variables and their interaction. As response variable, we used either a pairwise linear FST matrix [measured as FST/(1 − FST)] derived from 616 neutral SNPs or the allele frequency differential delta (AFD, Zhu et al. 2005) for each of the two outlier SNPs (here, we considered as candidate to divergent selection those SNPs detected as outliers by at least two distinct methods). As predictor variables, we used the GEO matrix to assess the effect of IBD, as well as CLIM, ELEV, and SOIL matrices to access the effect of IBE. To avoid multicollinearity, we calculated the variance inflation index and Pearson’s correlation coefficient among explanatory variables and considered VIF < 2 and correlation <0.7 as acceptable to include the variable in the model.
We confronted models including either GEO, CLIM, ELEV, or SOIL matrix as a single predictor and models combining two, three, or four predictors against a null model (see Table 4). The analysis was performed with the R package MCMCGLMM (Hadfield 2010), following scripts available in Lexer et al. (2014). MCMCGLMM was run with a burn-in of 500,000 followed by 2,000,000 MCMC iterations with a thinning interval of 750, under standard priors. We accounted for the lack of independence between pairs of populations by fitting a multiple membership model. Chain convergences were checked using the ‘CODA’ R package. We used the DIC to compare models and draw conclusions on the relative roles of IBD versus IBE in the divergence of P. lanuginosa within the Cerrado.
P ST–F ST comparisons
We also evaluated the relative importance of selection and neutral evolutionary processes in P. lanuginosa by comparing differentiation in quantitative ecophysiological traits (as measured by PST) and neutral markers derived from ddRAD sequencing (as measured by FST, Weir and Cockerham 1984) among a subset of eight P. lanuginosa populations from the Cerrado (Table 1). To represent the degree of phenotypic differentiation among these populations, we employed an approximation of QST called PST (Merilä and Crnokrak 2001). PST is directly calculated from the total phenotypic variance with no distinction between the relative contribution of genetic and environmental variation (Leinonen et al. 2006; Silva and Silva 2018) and has been applied to distinct biological systems as an alternative of QST (e.g., He et al. 2013; Shinn et al. 2015), including to systems exhibiting strong genetic structure (Miller et al. 2019; Pometti et al. 2019). We assessed trait values in a common garden; we used this approach because a known family structure would be required to estimate the additive genetic variance component of QST. The logic of PST–FST comparison relies on the assumption that FST values obtained from neutral molecular markers reflect the divergence induced by genetic drift without selection (Merilä and Crnokrak 2001). Thus, PST > FST or PST < FST mean that quantitative traits show a higher or lower level of differentiation than expected under the influence of genetic drift alone, as induced by divergent or balancing selection, respectively; while PST = FST means that no departure from neutral expectations can be detected (Merilä and Crnokrak 2001). This analysis assumes that mutation rates at coding genes and neutral regions of the genome are uniform or that time since divergence among populations is short enough or migration is high enough to disregard mutations (Leinonen et al. 2013).
Population pairwise PST was calculated from between-population (σ2B) and within population (σ2w) components of variance for each of the 11 ecophysiological traits following Brommer’s (2011) formula PST = cσ2B/(cσ2B + 2 h2σ2W) using R package ‘Pstat’ (Silva and Silva 2018). We assumed that the proportion of the total phenotypic variance which is due to additive genetic effects is equal to the narrow-sense heritability, as we have no previous data on contributions of additive genetic variance to among (c) and within (h2) population differences in ecophysiological traits for our sampled populations. Because of the known problem associated with the lack of accurate estimation of the parameters c and h2 for a set of traits (see Brommer 2011), we assessed the strength of PST–FST comparisons exploring the variation range of 0 < c/h2 ≤ 1 (sensitivity analysis) using ‘Pstat’. In its turn, pairwise FST values were calculated using the ‘populations’ module of STACKS for the subset of 616 putatively neutral SNPs, excluding the two outliers detected by at least two of the employed approaches for the Cerrado populations. Confidence intervals of PST and FST values were determined with the bootstrap method (1000 repetitions) under a confidence level of 0.95%. PST was considered to significantly differ from FST when their confidence intervals did not overlap. Finally, we used an analysis of variance (ANOVA), followed by a Tukey test, to compare means among multiple populations for each trait bearing a signature of selection using the software R.
Results
Genetic diversity and structure patterns
Three lanes of ddRAD-sequencing (40–48 samples per lane) using ION Torrent technology resulted in ~1,992,450 single end reads per P. lanuginosa sample, with an average read length of 178 bases and an average of 84% bases with Q score above 20 (Table S3). PYRAD assembled a total of 13,858 ddRAD-based SNP loci. After VCFTOOLS filtering following missing data criteria for loci and individuals, our final matrix included 115 individuals and 688 SNPs. Final matrices for the Cerrado and Yungas subsets in their turn had a total of 618 and 714 SNPs, and 95 and 21 individuals, respectively.
We found extremely low within-population genetic variation in P. lanuginosa (Table 2). The proportion of polymorphic sites per population ranged from 1.93 to 18.52, while nucleotide diversity calculated for the SNPs ranged from 0.0118 to 0.0885 (Table 2). Observed and expected heterozygosities per population varied from 0 to 0.0009 and 0.0092 to 0.0753, respectively (Table 2).
Pairwise FST and G′ST are generally high, with exception of neighbor populations SJC-YUR (Table S4). Independent analyses consistently recovered the same genetic structure for P. lanuginosa. The neighbor-net analysis revealed two major splits corresponding to Cerrado and Yungas populations with bootstrap support of 100%, despite Eastern Cerrado populations being slightly differentiated from those belonging to the Central-Western Cerrado distribution (Fig. 2). INSTRUCT clustered populations into three groups corresponding to Eastern Cerrado, Central-Western Cerrado and Yungas (east-west gradient of genetic variance), with low levels of admixture between both Cerrado clusters, and high selfing rates within clusters (0.956, 0.961, and 0.961, respectively; Fig. 1A, B). AMOVA showed that the largest part of the genomic variance is partitioned among groups, either when contrasting Cerrado and Yungas or Eastern Cerrado, Central-western Cerrado and Yungas (FCT = 0.54 and 0.53, respectively; p < 0.001), suggesting historical genetic isolation among these groups (Table 3). About 24% and 19% of the variation accounted for differences among populations nested within each of these groups, respectively, while 22% and 27% accounted for within-population variance (Table 3).
Drift and selection effects
BAYESCAN detected a total of 34 FST outliers (i.e., SNPs putatively under selection), while BAYPASS and PCADAPT identified 5 and 43 outlier SNPs, respectively, among all populations sampled (Fig. 3A). Nevertheless, only two SNPs were regarded as candidates for being under selection due to their detection by at least two distinct methods (Fig. 3A). Both SNPs are candidates for directional selection, as demonstrated by their higher differentiation in terms of FST in BAYESCAN analysis (Fig. S1A). When searching for FST outliers within the Cerrado distribution, BAYESCAN pointed to a total of 17 outliers, while BAYPASS and PCADAPT detected 4 and 59 outlier SNPs, respectively. For this data set, the same two outlier SNPs were recovered by at least two different methods (Fig. 3B), both putatively under directional selection (Fig. S1B, Table 4).
According to Mantel analyses, geographical distances are strongly correlated with climatic distances (r = 0.76, p < 0.001), and moderately correlated with other environmental distances derived from soil and elevation data (r = 0.26, p < 0.01; r = 0.27, p < 0.01; respectively; Table S5) within the Cerrado. As expected, climatic and elevation distances are also correlated with each other (r = 0.51, p < 0.01; Table S5). When using an FST matrix obtained from 616 neutral SNPs (Cerrado assembly), GLMM identified the model using soil as the single predictor (SOIL) as the best to predict genomic variance, but not significantly better (i.e., ΔDIC < 2) than the one combining soil and geographic pairwise distances (GEO + SOIL) (Table 4). GLMMs suggested an effect of both climatic and geographical distances on the AFD of the two outlier SNPs (Table 4). When contrasted to other predictors, the model that included all environmental distances and geographical distances as explanatory variables achieved the greatest DIC support (Table 4), indicating that genetic variation at these outlier loci is better explained by a combined effect of IBE and IBD. However, the full model was not significantly better than the model that included only climatic and geographical distances (GEO + CLIM; ΔDIC < 2) (Table 4).
The confidence intervals for most PST estimates derived from 11 ecophysiological traits overlapped with the confidence interval of FST among eight Cerrado populations (FST = 0.503, CI = 0.456–0.550; Fig. 4A), thus fitting the neutral expectation. Nevertheless, PST values were significantly higher than FST for two measured traits, the decay of both maximal photochemical efficiency (Fv/Fm) and light saturation coefficient (Ik) under drought stress (PST = 0.943 and 0.847; CI = 0.878–0.987 and 0.696–0.931, respectively; Fig. 4A), which may be caused by directional selection. Because the confidence interval of PST for Ik (mean PST = 0.758, CI = 0.551–0.943) nearly overlapped with that of FST, we did not consider it as different (Fig. 4A). Sensitivity analyses indicated that these results are robust to c/h2 variation, except for extremely low values of c/h2 (Fig. S2). ANOVAs showed that populations NAT and TIR are significantly less tolerant to drought stress in terms of Fv/Fm (Tukey test, p < 0.05; Fig. 4B), whereas RON is less tolerant to drought stress in terms of Ik (Fig. 4C).
Traits related to heat, light, and drought stresses are shown in red, green and blue, respectively (A). Boxplots comparing means among populations for each trait bearing a signature of divergent selection, i.e. decay of Fv/Fm (B) and Ik (C) under drought stress. CTmax = Upper critical thermal limit, z = sensitivity to temperature change, ETRmax = maximum relative electron transport rate, α = initial slope of the curve of light-response, Ik = light saturation coefficient, and Fv/Fm = maximal photochemical efficiency.
Discussion
The application of combined data analysis including high throughput sequencing and ecophysiological traits refined our understanding of the mechanisms underlying the population differentiation in the widespread tropical bromeliad P. lanuginosa. The analysis of hundreds of loci revealed low genetic variation within populations and pronounced structure among populations of this highly endogamous and patchily distributed species, a pattern that suggests a strong role of genetic drift. Our data also revealed few outlier loci and few phenotypic traits putatively under directional selection, which suggests an additional role of natural selection, despite strong drift, in populations subject to similar microhabitat conditions. Below we discuss these main findings based on information about the effects of drift and selection on population differentiation in this species and highlight which species’ ecological features may be particularly linked to the genomic patterns recovered here.
We found a remarkably low genetic diversity within P. lanuginosa populations using hundreds of SNPs, in agreement with our previous study based on eight microsatellites (Leal et al. 2019). In fact, such a low within-population genetic diversity is expected in a predominantly selfing species occurring in small and spatially isolated populations. Although both studies evidenced a remarkably low genetic variability, heterozygosities are even lower for the SNP data set than for microsatellites (Leal et al. 2019). Such differences are likely due to distinct mutation rates between makers, typically higher for microsatellites by several orders of magnitude (Chakraborty et al. 1997; Haasl and Payseur 2011). Furthermore, we cannot rule out the effect of lower population sample sizes used here. Despite these differences, our results indicate that the low levels of variability resulting from strong genetic drift may decrease the adaptive potential of P. lanuginosa to novel environmental conditions (Messer and Petrov 2013), but might not prevent populations to locally adapt. Accordingly, combined analyses detected a limited number of outlier SNPs (corresponding to 0.2–0.3% of the total) under putative directional selection in P. lanuginosa, highlighting the important role of genetic drift compared to selection in promoting population differentiation within non-continuously distributed organisms. Although we expect missing data to have some impact on the detection of outliers through inflating genetic differentiation parameters, missingness in the two candidate SNPs (ca. 30% of individuals) was unlikely to explain their higher FST values (FST = 0.902 and 0.903). Furthermore, these candidates SNPs were identified simultaneously by two methods that take into account different signatures left by natural selection at the molecular level (i.e., BAYESCAN and PCADAPT), suggesting thus a robust signature of putative directional selection.
Our study also showed that PST values among Cerrado populations for nine out 11 ecophysiological traits were within neutral expectations. Although we expect contrasting macroclimatic conditions across the species distribution, P. lanuginosa populations generally occur within riparian forests, and may, therefore, be exposed to similar selective pressures across this wide distribution. Indeed, local adaptation is expected to be less frequent between populations experiencing similar environmental conditions (reviewed by Hereford 2009). Therefore, the consistent association to a specialized microhabitat suggests that niche conservatism may be the major process leading to the divergence, even between intra-specific lineages occurring in the Cerrado and Yungas.
On the other hand, the results of GLMMs suggested the existence of IBE determined by climate variation or by the combined effect of climate, soil, and elevation, in addition to IBD within the Cerrado for two candidate SNPs which exhibited a signature of putative directional selection. Furthermore, two traits related to drought stress (i.e., the decay of Fv/Fm and Ik under drought stress) showed an increased phenotypic variation in our PST–FST comparisons among Cerrado populations that were robust to c/h2 variation. Indeed, populations from the Cerrado exhibit varying levels of drought tolerance as measured by such traits, suggesting local adaptation to differences in micro-habitats and/or microclimates. Although riparian forests are buffered from abrupt climatic changes, levels of disturbance due to flooding or intermittent drought likely vary among P. lanuginosa sites. Together, these pieces of evidence suggest that selection acting on a few traits could overcome the effects of drift associated with the low observed effective population sizes and high inbreeding in this species. Because populations inhabiting the Cerrado are submitted to seasonal variation of precipitation, such specific traits could be seen as preliminary candidates for local adaptation (see Pujol et al. 2008), but still demand further investigation to confirm their importance.
Caution must be taken on interpreting results on ecophysiological traits identified to be under divergent selection because low sample sizes and violated assumptions in estimating FST and PST might create bias for comparisons and lead to the detection of false positives (Miller et al. 2008; Edelaar et al. 2011; Leinonen et al. 2013). One assumption of such analysis is that mutation rates at coding genes underlying traits are comparable to those of neutral regions of the genome (Leinonen et al. 2013). This assumption might be supported when mutation rates are high relative to migration rates or much time has passed since population divergence (Edelaar et al. 2011; Leinonen et al. 2013). It has been argued, however, that the use of SNPs can provide a more reliable neutral baseline differentiation as they experience lower mutation rates, which are comparable to those of the loci underlying quantitative traits (Roach et al. 2010; Whitlock 2011). Indeed, SNP markers likely prevents upward bias (i.e., QST or PST > FST, Edelaar et al. 2011). Notwithstanding, interpretation of results from genome scans of SNPs requires caution, because finding candidates to divergent selection is particularly difficult when the average level of population differentiation is high (De Mita et al. 2013; Hoban et al. 2016; Lowry et al. 2017). Also, more candidates for selection could arise from outlier tests for data sets with a higher density of markers (e.g., derived from whole genome sequencing). For our model species, we conclude, thus, that both PST–FST comparisons and genome scans may have rendered conservative rather than liberal results regarding the existence of traits and loci under divergent selection. Further studies could provide more insights on the relative importance of local adaptation for the differentiation of P. lanuginosa populations.
Our data also revealed high levels of genetic differentiation among P. lanuginosa populations (FST > 0.70, p value = 0.0001). Our RAD-based estimates are in strong agreement with previous findings based on microsatellites markers (global FST = 0.71; Leal et al. 2019). However, studies have generally reported smaller FST for microsatellite-based estimates than SNPs-based (e.g., Elhaik 2012). Although RAD-seq can accurately infer genetic structure even with relaxed levels of missing data (see Hodel et al. 2017), we cannot rule out that the excess of missing data has led to biased FST (see Arnold et al. 2013; Gautier et al. 2013). When using G′ST, which is generally less affected by within-population variability than FST, we estimated lower levels of genetic differentiation with SNPs (global G′ST = 0.770) than with microsatellite markers (global G′ST = 0.861, as calculated from the dataset of Leal et al. 2019). Patterns of genetic structure recovered here are similar to what has been reported for organisms distributed across islands or island-like terrestrial habitats with RAD markers (e.g. Funk et al. 2016; Wang et al. 2017). P. lanuginosa occurs as small and isolated populations and presents a combined set of ecological characteristics that likely limit gene flow. The species shows limited seed dispersal, owing to the tiny seeds that lack any specific dispersal mechanism (Smith and Downs 1974), a reduced overlap of flowering phenology among populations, and high rates of self-fertilization, which decrease opportunities for both seed and pollen-mediated gene flow thereby promoting differentiation.
Although genetic differentiation among populations is generally high, analyses consistently assigned P. lanuginosa populations to three genetic clusters occurring in the Eastern Cerrado, Central-western Cerrado and Yungas. Such divergence within the species also agrees with that previously detected using Sanger sequencing and microsatellite markers (Leal et al., 2019) and has been commonly reported for other Cerrado plant species (Resende-Moreira et al. 2017; Buzzati et al. 2018; reviewed by Leal et al. 2016). Because there are no obvious historical or geographical barriers to gene flow explaining the concordance among ecologically distinct species, this Eastern-Western genetic structure pattern deserves special attention and could be better investigated under a comparative framework. For riparian plants, such as P. lanuginosa, the genetic structure could also be explained due to historical drainage re-organization events, as has been shown for species inhabiting the Asian mountains valleys (Yue et al. 2002; Zhang et al. 2011), a hypothesis to be further investigated.
Patterns of genomic diversity and structure recovered for P. lanuginosa are likely under a strong influence of the high rates of self-fertilization. Despite the predicted benefits of selfing for reproductive assurance when pollinators and/or mates are rare, particularly during colonization events, high rates of self-fertilization may limit the species’ evolutionary potential (Herlihy and Eckert 2002; Wright et al. 2013). Because the strength of selection scales to the effective population size (Kimura 1983), natural selection may be less efficient in selfing populations and, to an even greater extent, in recently founded populations experiencing selfing (Laenen et al. 2018). Indeed, studies have found that small populations are less likely to display local adaptation, but the influence of mating system on limiting adaptation is still controversial (see Leimu and Fischer 2008; Hereford 2010; Clo et al. 2019). Selfing could theoretically promote local adaptation if selfing populations experience less gene flow than outcrossing populations (Linhart and Grant 1996; Hereford 2010). In addition, theoretical studies have shown that selfing tends to increase the effect of selection by exposing alleles to homozygosity that can be more easily removed or selected (Charlesworth and Charlesworth 1987; De Mita et al. 2013). The influence of self-fertilization on the efficiency of selection seems therefore unpredictable, and further empirical studies on organisms performing predominant selfing may help to test some of the theoretical predictions (see Clo et al. 2019).
In summary, genomic analyses with a set of hundreds of loci generated by ddRAD sequencing, and additional data on ecophysiological variation at the population level, found support for genetic drift as the main evolutionary mechanism responsible for divergence across the wide yet patchy distribution of P. laniginosa, a highly self-fertilized species. Natural selection may have an important role, particularly at maintaining highly advantageous variants or eliminating deleterious mutations within populations and thus allowing the persistence in small patches of riparian forests. Indeed, our results suggested that selection might be the dominant force underlying the phenotypic divergence of traits related to drought stress, while neutral processes likely explain the divergence for other ecophysiological traits measured in this study. Determining to what degree differentiation among populations is caused by adaptive versus neutral processes provides important insights about population differentiation across wide distribution ranges. As demonstrated by an increasing number of studies employing genome-wide markers, patterns of genetic diversity and structure are rarely explained by a single evolutionary force (e.g, Orsini et al. 2013, Moura et al. 2014; Gaither et al. 2015). Rather, complex patterns commonly arise when employing a wide genomic coverage and natural selection is often assumed as a relevant force to shape particular traits, even for populations experiencing strong drift (e.g. Prentice et al. 2017; Wang et al. 2017). The balance between genetic drift and natural selection and the relative contribution of plasticity to environmental adaptation seem to be strictly dependent on the ecological context. For predominantly selfing species inhabiting isolated but relatively stable habitats, such as riparian forests, drift may be stronger than selection for most loci. But even these relatively stable habitats can be subject to flooding or intermittent drought disturbance, imposing conditions in which plants must adapt to the environment either through genetic adaptation or phenotypic plasticity.
Data availability
The PyRAD outputs generated from the de novo assembly of RAD-seq data, as well as ecophysiological data collected from the common garden experiment, are available on the Dryad Digital Repository https://doi.org/10.5061/dryad.zpc866t84. Raw reads have been deposited in the NCBI SRA database (BioProject ID: PRJNA723200; BioSample accessions: SRR14279592-SRR14279706).
References
[dataset]Leal BSS, Chaves CJN, Graciano VA, Boury C, Huacre LAP, Heuertz M, Palma-Silva C. 2021. RAD-seq data from Evidence of local adaptation despite strong drift in a Neotropical patchily distributed bromeliad; Dryad; https://doi.org/10.5061/dryad.zpc866t84
Agashe D, Falk JJ, Bolnick DI (2011) Effects of founding genetic variation on adaptation to a novel resource. Evolution: Int J Org Evolution 65:2481–2491
Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA (2016) Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet 17:81
Antonelli A, Ariza M, Albert J, Andermann T, Azevedo J, Bacon C et al. (2018) Conceptual and empirical advances in Neotropical biodiversity research. PeerJ 6:e5644
Arnold B, Corbett‐Detig RB, Hartl D, Bomblies K (2013) RAD seq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol Ecol 22:3179–3190
Bacon CD, Moraes M, Jaramillo C, Antonelli A (2017) Endemic palm species shed light on habitat shifts and the assembly of the Cerrado and Restinga floras. Mol Phylogenetics Evolution 110:127–133
Baranzelli MC, Cosacov A, Espíndola A, del Rosario Iglesias M, Chan LM, Johnson LA et al. (2018) Echoes of the whispering land: interacting roles of vicariance and selection in shaping the evolutionary divergence of two Calceolaria (Calceolariaceae) species from Patagonia and Malvinas/Falkland Islands. Evolut Ecol 32:287–314
Barrett RD, Schluter D (2008) Adaptation from standing genetic variation. Trends Ecol Evolution 23:38–44
Brancalion PH, Oliveira GC, Zucchi MI, Novello M, van Melis J, Zocchi SS et al. (2018) Phenotypic plasticity and local adaptation favor range expansion of a Neotropical palm. Ecol Evolution 8:7462–7475
Brommer JE (2011) Whither PST? The approximation of QST by PST in evolutionary and conservation biology. J Evolut Biol 24:1160–1168
Buzatti RSO, Pfeilsticker TR, Magalhaes RF, Bueno ML, Lemos-Filho JP, Lovato MB (2018) Genetic and historical colonization analyses of an endemic savanna tree, qualea grandiflora, reveal ancient connections between amazonian savannas and cerrado core. Frontiers in Plant Science 9:981
Cardoso VJM (2010) An adapted thermal-gradient block for the germination of photoblastic seeds. Braz Arch Biol Technol 53:1267–1277
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis toolset for population genomics. Mol Ecol 22:3124–3140
Chakraborty R, Kimmel M, Stivers DN, Davison LJ, Deka R (1997) Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc Natl Acad Sci 94:1041–1046
Charlesworth B (2009) Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet 10:195–205
Charlesworth D, Charlesworth B (1987) Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Syst 18:237–268
Chattopadhyay B, Garg KM, Ramakrishnan U (2014) Effect of diversity and missing data on genetic assignment with RAD-Seq markers. BMC Res Notes 7:841
Chaves CJN, Leal BSS, de Lemos-Filho JP (2018) How are endemic and widely distributed bromeliads responding to warming temperatures? A case study in a Brazilian hotspot. Flora 238:110–118
Clo J, Gay L, Ronfort J (2019) How does selfing affect the genetic variance of quantitative traits? An updated meta‐analysis on empirical results in angiosperm species. Evolution 73:1578–1590
Coyne JA, Orr HA (2004) Speciation. Boston, Sinauer Associates Inc.
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499–510
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NEESTIMATOR v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Eaton DA (2014) PyRAD: assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics 30:1844–1849
Ebel ER, da Costa JM, Sorenson MD, Hill RI, Briscoe AD, Willmott KR, Mullen SP (2015) Rapid diversification associated with ecological specialization in Neotropical Adelpha butterflies. Mol Ecol 24:2392–2405
Edelaar PIM, Burraco P, Gomez‐Mestre I (2011) Comparisons between QST and FST—how wrong have we been? Mol Ecol 20:4830–4839
Elhaik E (2012) Empirical distributions of FST from large-scale human polymorphism data. PloS ONE 7:e49837
Excoffier L, Lischer HE (2010) Arlequin suite ver. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Flood PJ, Hancock AM (2017) The genomic basis of adaptation in plants. Curr Opin Plant Biol 36:88–94
Foll M, Gaggiotti OE (2008) A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Funk WC, Lovich RE, Hohenlohe PA, Hofman CA, Morrison SA, Sillett TS et al. (2016) Adaptive divergence despite strong genetic drift: genomic analysis of the evolutionary mechanisms causing genetic differentiation in the island fox (Urocyon littoralis). Mol Ecol 25:2176–2194
Gaither MR, Bernal MA, Coleman RR, Bowen BW, Jones SA, Simison WB, Rocha LA (2015) Genomic signatures of geographic isolation and natural selection in coral reef fishes. Mol Ecol 24:1543–1557
Gao H, Williamson S, Bustamante CD (2007) An MCMC approach for joint inference of population structure and inbreeding rates from multi-locus genotype data. Genetics 176:1635–1651
Gautier M (2015) Genome‐wide scan for adaptive divergence and association with population‐specific covariates. Genetics 201:1555–1579
Gautier M, Gharbi K, Cezard T, Foucaud J, Kerdelhué C, Pudlo P et al. (2013) The effect of RAD allele dropout on the estimation of genetic variation within and between populations. Mol Ecol 22:3165–3178
Gayevskiy V, Klaere S, Knight S, Goddard MR (2014) ObStruct: a method to objectively analyse factors driving population structure using Bayesian ancestry profiles. PLoS ONE 9:e85196
Glémin S (2007) Mating systems and the efficacy of selection at the molecular level. Genetics 177:905–916
Godoy O, de Lemos-Filho JP, Valladares F (2011) Invasive species can handle higher leaf temperature under water stress than Mediterranean natives. Environ Exp Bot 71:207–214
Günther T, Coop G (2013) Robust identification of local adaptation from allele frequencies. Genetics 195:205–220
Haasl RJ, Payseur BA (2011) Multi-locus inference of population structure: a comparison between single nucleotide polymorphisms and microsatellites. Heredity 106:158–171
Hadfield JD (2010) MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw 33:1–22.
Harvey MG, Brumfield RT (2015) Genomic variation in a widespread Neotropical bird (Xenops minutus) reveals divergence, population expansion, and gene flow. Mol Phylogenetics Evolution 83:305–316
He Y, Li R, Wang J, Blanchet S, Lek S (2013) Morphological variation among wild populations of Chinese rare minnow (Gobiocypris rarus): Deciphering the role of evolutionary processes. Zool Sci 30:475–483
Hedrick PW (2007) A standardized genetic differentiation measure. Evolution 59:1633–1638
Hendry AP (2015) Key questions on the role of phenotypic plasticity in eco-evolutionary dynamics. J Heredity 107:25–4
Hengl T, Mendes de Jesus J, Heuvelink GBM, Ruiperez Gonzalez M, Kilibarda M et al. (2017) SoilGrids250m: global gridded soil information based on machine learning. PLoS ONE 12:e0169748
Hereford J (2009) A quantitative survey of local adaptation and fitness trade‐offs. Am Naturalist 173:579–588
Hereford J (2010) Does selfing or outcrossing promote local adaptation? Am J Bot 97:298–302
Herlihy CR, Eckert CG (2002) Genetic cost of reproductive assurance in a self-fertilizing plant. Nature 416:6878–320
Hijmans RJ, Cameron SE, Parra JL, Jones PJ, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas International Journal of Climatology 25:1965–1978
Hoban S, Kelley JL, Lotterhos KE, Antolin MF, Bradburd G, Lowry DB et al. (2016) Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am Naturalist 188:379–397
Hodel RG, Chen S, Payton AC, McDaniel SF, Soltis P, Soltis DE (2017) Adding loci improves phylogeographic resolution in red mangroves despite increased missing data: comparing microsatellites and RAD-Seq and investigating loci filtering. Sci Rep 7:17598
Huang H, Knowles LL (2016) Unforeseen consequences of excluding missing data from next-generation sequences: simulation study of RAD sequences. Syst Biol 65:357–365
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evolution 23:254–267
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) DiveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evolution 4:782–788
Kimura M (1983). The Neutral Theory of Molecular Evolution. Cambridge, Cambridge Univ Press.
Labouriau LG (1977) Thermal gradient block for germination experiments. Rev Brasileira de Biol 37:295–305
Laenen B, Tedder A, Nowak MD, Toräng P, Wunder J, Wötzel S et al. (2018). Demography and mating system shape the genome-wide impact of purifying selection in Arabis alpina. Proc Natl Acad Sci 115:816–821.
Lande R, Barrowclough G (1987) Effective population size, genetic variation, and their use in population management. In: Soulé, M. (ed.), Viable Populations for Conservation. Cambridge, Cambridge University Press, p. 87–124
Leal BSS, Pinheiro F, Palma-Silva C (2016) Phylogeographic studies depict the role of space and time scales of plant speciation in a highly diverse Neotropical region. Crit Rev Plant Sci 35:215–230
Leal BSS, Graciano VA, Chaves CJN, Huacre LAP, Heuertz M, Palma-Silva C (2019) Dispersal and local persistence shape the genetic structure of a widespread Neotropical plant species with a patchy distribution. Ann Bot 124:499–512
Leal BSS (2018). Filogeografia e genômica de populações de Pitcairnia lanuginosa (Bromeliaceae). Doctoral thesis. Universidade Estadual Paulista, Rio Claro, Brazil
Leimu R, Fischer M (2008) A meta‐analysis of local adaptation in plants. PLoS ONE 3:e4010
Leinonen T, Cano JM, Makinen H, Merilä J (2006) Contrasting patterns of body shape and neutral genetic divergence in marine and lake populations of threespine sticklebacks. J Evolut Biol 19:1803–1812
Leinonen T, McCairns RS, O’hara RB, Merilä J (2013) QST–FST comparisons: evolutionary and ecological insights from genomic heterogeneity. Nat Rev Genet 14:179–190
Lexer C, Wüest RO, Mangili S, Heuertz M, Stölting KN, Pearman PB et al. (2014) Genomics of the divergence continuum in an African plant biodiversity hotspot, I: drivers of population divergence in Restio capensis (Restionaceae). Mol Ecol 23:4373–4386
Linhart YB, Grant MC (1996) Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Syst 27:237–277
Lischer HE, Excoffier L (2012) PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28:298–299
Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, Storfer A (2017) Breaking RAD: An evaluation of the utility of restriction site-associated DNA sequencing for genome scans of adaptation. Mol Ecol Resour 17:142–152
Luu K, Bazin E, Blum MG (2017) pcadapt: an R package to perform genome scans for selection based on principal component analysis. Mol Ecol Resour 17:67–77
McCormack JE, Zellmer AJ, Knowles LL (2010) Does niche divergence accompany allopatric divergence in Aphelocoma jays as predicted under ecological speciation?: Insights from tests with niche models. Evolution 64:1231–1244
Merilä J, Crnokrak P (2001) Comparison of genetic differentiation at marker loci and quantitative traits. J Evolut Biol 14:892–903
Messer PW, Petrov DA (2013) Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol Evolution 28:659–669
Miller JR, Hamilton MB, Wood BP (2008) FST and QST under neutrality. Genetics 180:1023–1037
De Mita S, Thuillet AC, Gay L, Ahmadi N, Manel S, Ronfort J, Vigouroux Y (2013) Detecting selection along environmental gradients: analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol Ecol 22:1383–1399
Moura AE, Kenny JG, Chaudhuri R, Hughes MA, Welch J, Reisinger RR et al. (2014) Population genomics of the killer whale indicates ecotype evolution in sympatry involving both selection and drift. Mol Ecol 23:5179–5192
Nadeau NJ, Martin SH, Kozak KM, Salazar C, Dasmahapatra KK, Davey JW et al. (2013) Genome-wide patterns of divergence and gene flow across a butterfly radiation. Mol Ecol 22:814–826
Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA (2013) Genotyping-by-sequencing in ecological and conservation genomics. Mol Ecol 22:2841–2847
Nistelberger H, Byrne M, Coates DJ, Roberts D (2015) Genetic drift drives evolution in the bird-pollinated, terrestrial island endemic Grevillea georgeana (Proteaceae). Botanical J Linn Soc 178:155–168
Orsini L, Andrew R, Eizaguirre C (2013) Evolutionary ecological genomics. Mol Ecol 22:527–531
Orsini L, Mergeay J, Vanoverbeke J, De Meester L (2013) The role of selection in driving landscape genomic structure of the waterflea Daphnia magna. Mol Ecol 22:583–601
Ortego J, Aguirre MP, Cordero PJ (2010) Population genetics of Mioscirtus wagneri, a grasshopper showing a highly fragmented distribution. Mol Ecol 19:472–483
Palma-Silva C, Wendt T, Pinheiro F, Barbará T, Fay MF, Cozzolino S, Lexer C (2011) Sympatric bromeliad species (Pitcairnia spp.) facilitate tests of mechanisms involved in species cohesion and reproductive isolation in Neotropical inselbergs. Mol Ecol 20:3185–3201
Perrier C, Ferchaud AL, Sirois P, Thibault I, Bernatchez L (2017) Do genetic drift and accumulation of deleterious mutations preclude adaptation? Empirical investigation using RAD seq in a northern lacustrine fish. Mol Ecol 26:6317–6335
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135
Plummer M, Best N, Cowles K, Vines K (2006) CODA: Convergence Diagnosis and Output Analysis for MCMC. R N. 6:7–11
Pometti CL, Bessega CF, Cialdella AM, Ewens M, Saidman BO, Vilardi JC (2019) Evidence of local adaptation and stabilizing selection on quantitative traits in populations of the multipurpose American species Acacia aroma (Fabaceae). Botanical J Linn Soc 191:128–141
Prentice MB, Bowman J, Khidas K, Koen EL, Row JR, Murray DL, Wilson PJ (2017) Selection and drift influence genetic differentiation of insular Canada lynx (Lynx canadensis) on Newfoundland and Cape Breton Island. Ecol Evolution 7:3281–3294
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pujol B, Wilson AJ, Ross RIC, Pannell JR (2008) Are QST–FST comparisons for natural populations meaningful? Mol Ecol 17:4782–4785
Resende-Moreira LC, de Vasconcelos PN, Souto AP, Menezes APA, de Lemos-Filho JP, Lovato MB (2017) East-west divergence in central Brazilian Cerrado revealed by cpDNA sequences of a bird-dispersed tree species. Biochemical Syst Ecol 70:247–253
Rezende EL, Castañeda LE, Santos M (2014) Tolerance landscapes in thermal ecology. Funct Ecol 28:799–809
Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT et al. (2010) Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328:636–639
Savolainen O, Lascoux M, Merila J (2013) Ecological genomics of local adaptation. Nat Rev Genet 14:807–820
Shinn C, Blanchet S, Loot G, Lek S, Grenouillet G (2015) Phenotypic variation as an indicator of pesticide stress in gudgeon: accounting for confounding factors in the wild. Sci Total Environ 538:733–752.
Silva SB, Silva A (2018) Pstat: an R package to assess population differentiation in phenotypic traits. R J 10:447–454
Smith LB, Downs RJ (1974) Pitcairnioideae (Bromeliaceae). Flora Neotropica Monogr 14:1–658
Sobel JM, Chen GF, Watt LR, Schemske DW (2010) The biology of speciation. Evolution 64:295–315
Vieira EA, Silva KR, Oriani A, Moro CF, Braga MR (2017b) Mechanisms of desiccation tolerance in the bromeliad Pitcairnia burchellii Mez: biochemical adjustments and structural changes. Plant Physiol Biochem 121:21–30
Vieira EA, Centeno DC, Freschi L, Silva EA, Braga MR (2017a) The dual strategy of the bromeliad Pitcairnia burchellii Mez to cope with desiccation. Environ Exp Bot 143:135–148
Wang J, Feng C, Jiao T, Von Wettberg EB, Kang M (2017) Genomic signature of adaptive divergence despite strong nonadaptive forces on edaphic islands: a case study of Primulina juliae. Genome Biol Evolution 9:3495–3508
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358
Whitlock MC (2011) GST and D do not replace FST. Mol Ecol 20:1083–1091
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Wright SI, Kalisz S, Slotte T (2013) Evolutionary consequences of self-fertilization in plants. Proc R Soc B 280:20130133
Yoichi W, Kawamata I, Matsuki Y, Suyama Y, Uehara K, Ito M (2018) Phylogeographic analysis suggests two origins for the riparian azalea Rhododendron indicum (L.) Sweet. Heredity 121:594–604
Yue L-L, Chen G, Sun W-B, Sun H (2002) Phylogeography of Buddleja crispa (Buddlejaceae) and its correlation with drainage system evolution in southwestern China. Am J Bot 99:1726–1735
Zhang T-C, Comes HP, Sun H (2011) Chloroplast phylogeography of Terminalia franchetii (Combretaceae) from the eastern Sino-Himalayan region and its correlation with historical river capture events. Mol Phylogenetics Evolution 60:1–12
Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T et al. (2005) Admixture mapping for hypertension loci with genome-scan markers. Nat Genet 37:177–181
Acknowledgements
We thank S. Nazareno, M. Arakaki, and P.H. Egoavil for assisting in sampling and permission procedures, and E. Guichoux for helping in the preparation of ddRAD-seq libraries. We are also grateful to K. Holsinger and two anonymous reviewers for providing helpful comments on the earlier version of the manuscript and to the Genotoul bioinformatics platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) and the Center for Scientific Computing (NCC/GridUNESP) of the São Paulo State University (UNESP) for providing computing and storage resources. This work was supported by Fundação de Apoio à Pesquisa do Estado de São Paulo (FAPESP; 2014/15588-6) and has benefited from the support of a grant from Investissement d’Avenir grants of the ANR (CEBA:ANR-10-LABX-25-01). BSSL and CJNC received scholarships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES; code 001) and FAPESP (2014/08087-0 and 2016/04396-4). BSSL also received a LabEx COTE mobility grant and a BEPE FAPESP scholarship (2016/20273-0). CPS was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; 300819/2016-1). Collection and export permits were granted by SISBIO (no. 44062-1), SERFOR (RDG no. 2017-2016), IDEFLOR-Bio/PA (no. 001/15), SEMARH/GO (no. 187/2014) and IEF/MG (no. 081/2014).
Author information
Authors and Affiliations
Contributions
BSSL, CJNC, MH, and CPS conceived the ideas; BSSL, CJNC, LAPH, and CPS sampled the populations; BSSL, VAG, and CB collected molecular data; CJNC measured the ecophysiological traits; BSSL and CJNC analyzed the data; and BSSL, CJNC, MH, and CPS led the writing. All authors read and accepted the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Thomas Meagher
Supplementary information
Rights and permissions
About this article
Cite this article
Leal, B.S.S., Chaves, C.J.N., Graciano, V.A. et al. Evidence of local adaptation despite strong drift in a Neotropical patchily distributed bromeliad. Heredity 127, 203–218 (2021). https://doi.org/10.1038/s41437-021-00442-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-021-00442-9
This article is cited by
-
Genetic diversity and population structure of Aechmea distichantha (Bromeliaceae), a widely geographically distributed species in South America
Plant Systematics and Evolution (2023)