Abstract
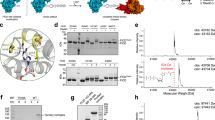
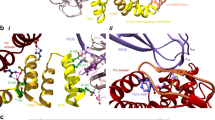
Fic (filamentation induced by cAMP) proteins regulate diverse cell signaling events by post-translationally modifying their protein targets, predominantly by the addition of an AMP (adenosine monophosphate). This modification is called Fic-mediated adenylylation or AMPylation. We previously reported that the human Fic protein, HYPE/FicD, is a novel regulator of the unfolded protein response (UPR) that maintains homeostasis in the endoplasmic reticulum (ER) in response to stress from misfolded proteins. Specifically, HYPE regulates UPR by adenylylating the ER chaperone, BiP/GRP78, which serves as a sentinel for UPR activation. Maintaining ER homeostasis is critical for determining cell fate, thus highlighting the importance of the HYPE-BiP interaction. Here, we study the kinetic and structural parameters that determine the HYPE-BiP interaction. By measuring the binding and kinetic efficiencies of HYPE in its activated (Adenylylation-competent) and wild type (de-AMPylation-competent) forms for BiP in its wild type and ATP-bound conformations, we determine that HYPE displays a nearly identical preference for the wild type and ATP-bound forms of BiP in vitro and preferentially de-AMPylates the wild type form of adenylylated BiP. We also show that AMPylation at BiP’s Thr366 versus Thr518 sites differentially affect its ATPase activity, and that HYPE does not adenylylate UPR accessory proteins like J-protein ERdJ6. Using molecular docking models, we explain how HYPE is able to adenylylate Thr366 and Thr518 sites in vitro. While a physiological role for AMPylation at both the Thr366 and Thr518 sites has been reported, our molecular docking model supports Thr518 as the structurally preferred modification site. This is the first such analysis of the HYPE-BiP interaction and offers critical insights into substrate specificity and target recognition.








Similar content being viewed by others
Abbreviations
- Fic:
-
filamentation induced by cAMP
- AMP:
-
adenosine monophosphate
- HYPE:
-
Huntingtin yeast interacting protein E
- UPR:
-
unfolded protein response
- ER:
-
endoplasmic reticulum
- GS:
-
glutamine synthetase
- GS-ATase:
-
glutamine synthetase adenylyltranferase
- BiP:
-
binding immunoglobulin protein
- NBD:
-
nucleotide binding domain
- SBD:
-
substrate binding domain
References
Allan RK, Ratajczak T (2011) Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16:353–367
Amin-Wetzel N, Saunders RA, Kamphius MJ, Rato C, Preissler S, Harding HP, Ron D (2017) Cell 171:1625–1637
Broncel M, Serwa RA, Bunney TD, Katan M, Tate EW (2016) Global Profiling of Huntingtin-associated protein E (HYPE)-Mediated AMPylation through a Chemical Proteomic Approach. Mol Cell Proteomics 15(2):715–725. https://doi.org/10.1074/mcp.O115.054429
Bunney TD, Cole AR, Broncel M, Esposito D, Tate EW, Katan M (2014) Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions. Structure 22:1831–1843
Casey AK, Orth K (2017) Enzymes involved in AMPylation and deAMPylation. Chem Rev 118:1199–1215
Casey AK, Moehlman AT, Zhang J, Servage KA, Kramer H, Orth K (2017) Fic-mediated deAMPylation is not dependent on homodimerization and rescues toxic AMPylation in flies. J Biol Chem 292:21193–21204
Castro-Roa D, Garcia-Pino A, De Gieter S, van Nuland NAJ, Loris R, Zenkin N (2013) The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol 9:811–817
Chambers JE, Petrova K, Tomba G, Vendruscolo M, Ron D (2012) ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J Cell Biol 198:371–385
Cruz JW, Rothenbacher FP, Maehigashi T, Lane WS, Dunham CM, Woychik NA (2014) Doc toxin is a kinase that inactivates elongation factor Tu. J Biol Chem 289:7788–7798
Dietz N, Huber M, Sorg I, Goepfert A, Harms A, Schirmer T, Dehio C (2021) Structural basis for selective AMPylation of Rac-subfamily GTPases by Bartonella effector protein 1 (Bep1). Proc Natl Acad Sci U S A 118:12. https://doi.org/10.1073/pnas.2023245118
Engel P, Goepfert A, Stanger FV, Harms A, Schmidt A, Schirmer T, Dehio C (2012) Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 482:107–U138
Faber PW, Barnes GT, Srinidhi J, Chen JM, Gusella JF, MacDonald ME (1998) Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet 7:1463–1474
Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou J-M (2012) A Xanthomonas uridine 5'-monophosphate transferase inhibits plant immune kinases. Nature 485:114–U149
Haas IG (1994) BIP (GRP78), An essential Hsp70 resident protein in the endoplasmic-reticulum. Experientia 50:1012–1020
Ham H, Woolery AR, Tracy C, Stenesen D, Kraemer H, Orth K (2014) Unfolded protein response-regulated Drosophila Fic (dFic) protein reversibly AMPylates BiP Chaperone during endoplasmic reticulum homeostasis. J Biol Chem 289:36059–36069
Hendershot LM, Ting J, Lee AS (1988) Identity of the immunoglobulin heavy-chain-binding protein with the 78,000-dalton glucose-regulated protein and the role of posttranslational modifications in its binding function. Mol Cell Biol 8:4250–4256
Kinch LN, Yarbrough ML, Orth K, Grishin NV (2009) Fido, a Novel AMPylation Domain Common to Fic, Doc, and AvrB. PLoS One 4:e5818
Kingdon HS, Shapiro BM, Stadtman ER (1967) Regulation of glutamine synthetase.8. Atp - glutamine synthetase adenylyltransferase an enzyme that catalyzes alterations in regulatory properties of glutamine synthetase. Proc Natl Acad Sci U S A 58:1703–1710
Mattoo S, Durrant E, Chen MJ, Xiao J, Lazar CS, Manning G, Dixon JE, Worby CA (2011) Comparative analysis of Histophilus somni immunoglobulin-binding protein A (IbpA) with other Fic domain-containing enzymes reveals differences in substrate and nucleotide specificities. J Biol Chem 286:32834–32842
Moehlman AT, Casey AK, Servage K, Orth K, Kramer H (2018) Adaptation to constant light requires Fic-mediated AMPylation of BiP to protect against reversible photoreceptor degeneration. ELife 7:e38752
Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR (2011) Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477:103–U122
Nikita P, Truman C, A.W., and Truttmann, M.C. (2020) Post-translational modifications of Hsp70 family proteins: expanding the chaperone code. J Biol Chem 295(31):10689–10708
Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M (2010) Quantitative Phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3:ra3
Perera LA, Rato C, Yan Y, Neidhardt L, McLaughlin SH, Read RJ, Preissler S, Ron D (2019) An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP. EMBO J 38(21):e102177. https://doi.org/10.15252/embj.2019102177
Peterson LX, Kim H, Esquivel-Rodriguez J, Roy A, Han XS, Shin WH, Zhang J, Terashi G, Lee M, Kihara D (2017a) Human and server docking prediction for CAPRI round 30-35 using LZerD with combined scoring functions. Proteins-Structure Function and Bioinformatics 85:513–527
Peterson L, Jamroz M, Kolinski A, Kihara D (2017b) Predicting real-valued protein residue fluctuation using FlexPred. in Prediction of Protein Secondary Structure (Zhou, Y., Kloczkowski, A., Faraggi, E., and Yang, Y. eds.) 175-186
Preissler S, Rato C, Chen R, Antrobus R, Ding S, Fearnley IM, Ron D (2015) AMPylation matches BiP activity to client protein load in the endoplasmic reticulum. Elife 4
Preissler S, Rohland L, Yan Y, Chen R, Read RJ, Ron D (2017a) AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional activation. ELife 6:e29428
Preissler S, Rato C, Perera LA, Saudek V, Ron D (2017b) FICD acts bifunctionally to AMPylate and de-AMPylate the endoplasmic reticulum chaperone BiP. Nat Struct Mol Biol 24:23–29
Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8:519–529
Sanyal A, Chen AJ, Nakayasu ES, Lazar CS, Zbornik EA, Worby CA, Koller A, Mattoo S (2015) A Novel Link between Fic (Filamentation Induced by cAMP)-mediated Adenylylation/AMPylation and the Unfolded Protein Response. J Biol Chem 290:8482–8499
Sengupta R, Poderycki MJ, Mattoo S (2019) CryoAPEX - an electron tomography tool for subcellular localization of membrane proteins. J Cell Sci 132(6):jcs222315. https://doi.org/10.1242/jcs.222315
Shapiro BM, Kingdon HS, Stadtman ER (1967) Regulation Of Glutamine Synthetase .7. Adenylyl glutamine synthetase - a new form of enzyme with altered regulatory and kinetic properties. Proc Natl Acad Sci U S A 58:642
Sreelatha A, Yee SS, Lopez VA, Park BC, Kinch LN, Pilch S, Servage KA, Zhang J, Jiou J, Karasiewicz-Urbańska M, Łobocka M, Grishin NV, Orth K, Kucharczyk R, Pawłowski K, Tomchick DR, Tagliabracci VS (2018) Protein AMPylation by an evolutionarily conserves pseudokinase. Cell 175:809–821
Tan Y, Arnold RJ, Luo Z-Q (2011) Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci U S A 108:21212–21217
Truttmann MC, Cruz VE, Guo XZ, Engert C, Schwartz TU, Ploegh HL (2016) The Caenorhabditis elegans protein FIC-1 is an AMPylase that covalently modifies heat-shock 70 family proteins, translation elongation factors and histones. PLoS Genet 12:e1006023
Venkatraman V, Yang YFD, Sael L, Kihara D (2009a) Protein-protein docking using region-based 3D Zernike descriptors. Bmc Bioinformatics 10
Venkatraman V, Sael L, Kihara D (2009b) Potential for protein surface shape analysis using spherical harmonics and 3D Zernike descriptors. Cell Biochem Biophys 54:23–32
Worby CA, Mattoo S, Kruger RP, Corbeil LB, Koller A, Mendez JC, Zekarias B, Lazar C, Dixon JE (2009) The Fic domain: regulation of cell signaling by adenylylation. Mol Cell 34:93–103
Wu S, Hong L, Wang Y, Yu J, Yang J, Yang J, Zhang H, Perrett S (2020) Kinetics of the conformational cycle of Hsp70 reveals the importance of the dynamic and heterogeneous nature of Hsp70 for its function. Proc Natl Acad Sci U S A 117(14):7814–7823. https://doi.org/10.1073/pnas.1914376117
Xiao J, Worby CA, Mattoo S, Sankaran B, Dixon JE (2010) Structural basis of Fic-mediated adenylylation. Nat Struct Mol Biol 17:1004–U1119
Yang J, Nune M, Zong YN, Zhou L, Liu QL (2015) Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure 23:2191–2203
Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K (2009) AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323:269–272
Acknowledgements
We thank Dr. Clint Chapple for advice on the design of kinetic experiments. We also thank the Purdue University office of Radiological and Environmental Management (REM) and Dr. Nicholas Carpita for access to the Liquid Scintillation Counter. Wild type HYPEΔ102/pSMT3 was a gift from Dr. Junyu Xiao and ERdJ6/pGEX-4T1 was kindly provided by Dr. David Ron. We also thank Dr. Linda Hendershot for additional J protein constructs and helpful discussions. Finally, we are grateful to members of the Mattoo lab for their helpful discussions.
Funding
This work was funded in part by the National Institute of General Medical Sciences of the National Institute of Health (R01GM10092), an Indiana Clinical and Translational Research Grant (CTSI-106564), and a Purdue Institute for Inflammation, Immunology, and Infectious Disease Core Start Grant (PI4D-209263) to SM; the National Institute of General Medical Sciences of the National Institutes of Health (R01GM123055) and the National Science Foundation (IOS1127027, DMS1614777) to DK; and grants from the Purdue Research Foundation (PRF-209104) and the Cancer Prevention Internship Program to AS.
Author information
Authors and Affiliations
Contributions
S.M. and A.S. conceived and designed the study. A.S., E.A.Z., B.G.W., and C.C. conducted the experiments. J.M. trained and provided advice on the BLI experiments. D.K. supervised the molecular docking studies. S.M. supervised the overall study. S.M. and A.S. wrote the manuscript. All authors revised and agreed to the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sanyal, A., Zbornik, E.A., Watson, B.G. et al. Kinetic and structural parameters governing Fic-mediated adenylylation/AMPylation of the Hsp70 chaperone, BiP/GRP78. Cell Stress and Chaperones 26, 639–656 (2021). https://doi.org/10.1007/s12192-021-01208-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-021-01208-2