Abstract
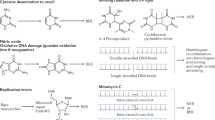
The GO system is part of the DNA base excision repair pathway and is required for the error-free repair of 8-oxoguanine (oxoG), one of the most common oxidative DNA lesions. Due to the ability of oxoG to form oxoG:A mispairs, this base is highly mutagenic. Its repair requires the action of two enzymes: 8-oxoguanine DNA glycosylase (Fpg or MutM in bacteria and OGG1 in eukaryotes), which removes oxoG from oxoG:C pairs, and adenine DNA glycosylase (MutY in bacteria and MUTYH in eukaryotes), which removes A from oxoG:A mispairs to prevent mutations. The third enzyme of the system (MutT in bacteria and MTH1 or NUDT1 in eukaryotes) hydrolyzes 8-oxo-2′-deoxyguanosine triphosphate, thus preventing its incorporation into DNA. Recent data point to the proteins of the GO system as promising targets for the therapy of cancer, autoimmune diseases, and bacterial infections. This review highlights the structure and specificity of the GO system in bacteria and eukaryotes and its unique role in mutation avoidance.




Similar content being viewed by others
REFERENCES
Staerck C., Gastebois A., Vandeputte P., Calenda A., Larcher G., Gillmann L., Papon N., Bouchara J.-P., Fleury M.J.J. 2017. Microbial antioxidant defense enzymes. Microb. Pathog. 110, 56–65.
Zharkov D.O. 2008. Base excision DNA repair. Cell. Mol. Life Sci. 65, 1544–1565.
van Loon B., Markkanen E., Hübscher U. 2010. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst.). 9, 604–616.
Shibutani S., Takeshita M., Grollman A.P. 1991. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 349, 431–434.
Moriya M. 1993. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-Oxoguanine in DNA induces targeted G ⋅ C→T ⋅ A transversions in simian kidney cells. Proc. Natl. Acad. Sci. U. S. A. 90, 1122–1126.
Michaels M.L., Cruz C., Grollman A.P., Miller J.H. 1992. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. U. S. A. 89, 7022–7025.
Michaels M.L., Tchou J., Grollman A.P., Miller J.H. 1992. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 31, 10964–10968.
Gad H., Koolmeister T., Jemth A.-S., Eshtad S., Jacques S.A., Ström C.E., Svensson L.M., Schultz N., Lundbäck T., Einarsdottir B.O., Saleh A., Göktürk C., Baranczewski P., Svensson R., Berntsson R.P.-A., et al. 2014. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 508, 215–221.
Huber K.V.M., Salah E., Radic B., Gridling M., Elkins J.M., Stukalov A., Jemth A.-S., Göktürk C., Sanjiv K., Strömberg K., Pham T., Warpman Berglund U., Colinge J., Bennett K.L., Loizou J.I., et al. 2014. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 508, 222–227.
Visnes T., Cázares-Körner A., Hao W., Wallner O., Masuyer G., Loseva O., Mortusewicz O., Wiita E., Sarno A., Manoilov A., Astorga-Wells J., Jemth A.-S., Pan L., Sanjiv K., Karsten S., et al. 2018. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science. 362, 834–839.
Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 130, 797–810.
Nguyen D., Joshi-Datar A., Lepine F., Bauerle E., Olakanmi O., Beer K., McKay G., Siehnel R., Schafhauser J., Wang Y., Britigan B.E., Singh P.K. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 334, 982–986.
Foti J.J., Devadoss B., Winkler J.A., Collins J.J., Walker G.C. 2012. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 336, 315–319.
Tchou J., Kasai H., Shibutani S., Chung M.-H., Laval J., Grollman A.P., Nishimura S. 1991. 8-oxoguanine (8‑hydroxyguanine. DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. U. S. A. 88, 4690–4694.
Tchou J., Bodepudi V., Shibutani S., Antoshechkin I., Miller J., Grollman A.P., Johnson F. 1994. Substrate specificity of Fpg protein: Recognition and cleavage of oxidatively damaged DNA. J. Biol. Chem. 269, 15318–15324.
Karakaya A., Jaruga P., Bohr V.A., Grollman A.P., Dizdaroglu M. 1997. Kinetics of excision of purine lesions from DNA by Escherichia coli Fpg protein. Nucleic Acids Res. 25, 474–479.
Bulychev N.V., Varaprasad C.V., Dormán G., Miller J.H., Eisenberg M., Grollman A.P., Johnson F. 1996. Substrate specificity of Escherichia coli MutY protein. Biochemistry. 35, 13147–13156.
Maki H., Sekiguchi M. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 355, 273–275.
Yudkina A.V., Shilkin E.S., Endutkin A.V., Makarova A.V., Zharkov D.O. 2019. Reading and misreading 8-oxoguanine, a paradigmatic ambiguous nucleobase. Crystals. 9, 269.
Tajiri T., Maki H., Sekiguchi M. 1995. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res. 336, 257–267.
Foster P.L., Lee H., Popodi E., Townes J.P., Tang H. 2015. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 112, E5990–E5999.
Wielgoss S., Barrick J.E., Tenaillon O., Wiser M.J., Dittmar W.J., Cruveiller S., Chane-Woon-Ming B., Médigue C., Lenski R.E., Schneider D. 2013. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. U. S. A. 110, 222–227.
Schalow B.J., Courcelle C.T., Courcelle J. 2011. Escherichia coli Fpg glycosylase is nonrendundant and required for the rapid global repair of oxidized purine and pyrimidine damage in vivo. J. Mol. Biol. 410, 183–193.
Auffret van der Kemp P., Thomas D., Barbey R., de Oliveira R., Boiteux S. 1996. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. U. S. A. 93, 5197–5202.
Nash H.M., Bruner S.D., Schärer O.D., Kawate T., Addona T.A., Spooner E., Lane W.S., Verdine G.L. 1996. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 6, 968–980.
Rosenquist T.A., Zharkov D.O., Grollman A.P. 1997. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. U. S. A. 94, 7429–7434.
Radicella J.P., Dherin C., Desmaze C., Fox M.S., Boiteux S. 1997. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 94, 8010–8015.
Roldán-Arjona T., Wei Y.-F., Carter K.C., Klungland A., Anselmino C., Wang R.-P., Augustus M., Lindahl T. 1997. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. U. S. A. 94, 8016–8020.
Bjørås M., Luna L., Johnsen B., Hoff E., Haug T., Rognes T., Seeberg E. 1997. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 16, 6314–6322.
Dherin C., Radicella J.P., Dizdaroglu M., Boiteux S. 1999. Excision of oxidatively damaged DNA bases by the human α-hOgg1 protein and the polymorphic α‑hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 27, 4001–4007.
Krishnamurthy N., Haraguchi K., Greenberg M.M., David S.S. 2008. Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry. 47, 1043–1050.
Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D.E. 1999. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U. S. A. 96, 13300–13305.
Minowa O., Arai T., Hirano M., Monden Y., Nakai S., Fukuda M., Itoh M., Takano H., Hippou Y., Aburatani H., Masumura K.-i., Nohmi T., Nishimura S., Noda T. 2000. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. U. S. A. 97, 4156–4161.
Sakumi K., Tominaga Y., Furuichi M., Xu P., Tsuzuki T., Sekiguchi M., Nakabeppu Y. 2003. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 63, 902–905.
Jaruga P., Xiao Y., Vartanian V., Lloyd R.S., Dizdaroglu M. 2010. Evidence for the involvement of DNA repair enzyme NEIL1 in nucleotide excision repair of (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosines. Biochemistry. 49, 1053–1055.
Sakumi K., Furuichi M., Tsuzuki T., Kakuma T., Kawabata S.-i., Maki H., Sekiguchi M. 1993. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 268, 23524–23530.
Slupska M.M., Baikalov C., Luther W.M., Chiang J.-H., Wei Y.-F., Miller J.H. 1996. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J. Bacteriol. 178, 3885–3892.
Hayakawa H., Taketomi A., Sakumi K., Kuwano M., Sekiguchi M. 1995. Generation and elimination of 8‑oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphate, a mutagenic substrate for DNA synthesis, in human cells. Biochemistry. 34, 89–95.
Pope M.A., David S.S. 2005. DNA damage recognition and repair by the murine MutY homologue. DNA Repair (Amst.). 4, 91–102.
Kundu S., Brinkmeyer M.K., Livingston A.L., David S.S. 2009. Adenine removal activity and bacterial complementation with the human MutY homologue (MUTYH) and Y165C, G382D, P391L and Q324R variants associated with colorectal cancer. DNA Repair (Amst.). 8, 1400–1410.
Russo M.T., De Luca G., Degan P., Parlanti E., Dogliotti E., Barnes D.E., Lindahl T., Yang H., Miller J.H., Bignami M. 2004. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res. 64, 4411–4414.
Xie Y., Yang H., Cunanan C., Okamoto K., Shibata D., Pan J., Barnes D.E., Lindahl T., McIlhatton M., Fishel R., Miller J.H. 2004. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 64, 3096–3102.
Ohno M., Sakumi K., Fukumura R., Furuichi M., Iwasaki Y., Hokama M., Ikemura T., Tsuzuki T., Gondo Y., Nakabeppu Y. 2014. 8-Oxoguanine causes spontaneous de novo germline mutations in mice. Sci. Rep. 4, 4689.
Pilati C., Shinde J., Alexandrov L.B., Assié G., André T., Hélias-Rodzewicz Z., Ducoudray R., Le Corre D., Zucman-Rossi J., Emile J.-F., Bertherat J., Letouzé E., Laurent-Puig P. 2017. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J. Pathol. 242, 10–15.
Boiteux S., Coste F., Castaing B. 2017. Repair of 8‑oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases. Free Radic. Biol. Med. 107, 179–201.
Fromme J.C., Verdine G.L. 2003. DNA lesion recognition by the bacterial repair enzyme MutM. J. Biol. Chem. 278, 51543–51548.
Bruner S.D., Norman D.P.G., Verdine G.L. 2000. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 403, 859–866.
Fromme J.C., Banerjee A., Huang S.J., Verdine G.L. 2004. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 427, 652–656.
Nakamura T., Meshitsuka S., Kitagawa S., Abe N., Yamada J., Ishino T., Nakano H., Tsuzuki T., Doi T., Kobayashi Y., Fujii S., Sekiguchi M., Yamagata Y. 2010. Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base. J. Biol. Chem. 285, 444–452.
Waz S., Nakamura T., Hirata K., Koga-Ogawa Y., Chirifu M., Arimori T., Tamada T., Ikemizu S., Nakabeppu Y., Yamagata Y. 2017. Structural and kinetic studies of the human Nudix hydrolase MTH1 reveal the mechanism for its broad substrate specificity. J. Biol. Chem. 292, 2785–2794.
Boon E.M., Pope M.A., Williams S.D., David S.S., Barton J.K. 2002. DNA-mediated charge transport as a probe of MutY/DNA interaction. Biochemistry. 41, 8464–8470.
Boon E.M., Livingston A.L., Chmiel N.H., David S.S., Barton J.K. 2003. DNA-mediated charge transport for DNA repair. Proc. Natl. Acad. Sci. U. S. A. 100, 12543–12547.
Boal A.K., Yavin E., Lukianova O.A., O’Shea V.L., David S.S., Barton J.K. 2005. DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry. 44, 8397–8407.
Yavin E., Boal A.K., Stemp E.D.A., Boon E.M., Livingston A.L., O’Shea V.L., David S.S., Barton J.K. 2005. Protein–DNA charge transport: Redox activation of a DNA repair protein by guanine radical. Proc. Natl. Acad. Sci. U. S. A. 102, 3546–3551.
Yavin E., Stemp E.D.A., O’Shea V.L., David S.S., Barton J.K. 2006. Electron trap for DNA-bound repair enzymes: A strategy for DNA-mediated signaling. Proc. Natl. Acad. Sci. U. S. A. 103, 3610–3614.
Maga G., Villani G., Crespan E., Wimmer U., Ferrari E., Bertocci B., Hübscher U. 2007. 8-Oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 447, 606–608.
Fazlieva R., Spittle C.S., Morrissey D., Hayashi H., Yan H., Matsumoto Y. 2009. Proofreading exonuclease activity of human DNA polymerase δ and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 37, 2854–2866.
Locatelli G.A., Pospiech H., Tanguy Le Gac N., van Loon B., Hubscher U., Parkkinen S., Syväoja J.E., Villani G. 2010. Effect of 8-oxoguanine and abasic site DNA lesions on in vitro elongation by human DNA polymerase ε in the presence of replication protein A and proliferating-cell nuclear antigen. Biochem. J. 429, 573–582.
Markkanen E., Castrec B., Villani G., Hübscher U. 2012. A switch between DNA polymerases δ and λ promotes error-free bypass of 8-oxo-G lesions. Proc. Natl. Acad. Sci. U. S. A. 109, 20401–20406.
Picher A.J., Blanco L. 2007. Human DNA polymerase lambda is a proficient extender of primer ends paired to 7,8-dihydro-8-oxoguanine. DNA Repair (Amst.). 6, 1749–1756.
Burak M.J., Guja K.E., Hambardjieva E., Derkunt B., Garcia-Diaz M. 2016. A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8‑oxo-dG. EMBO J. 35, 2045–2059.
van Loon B., Hübscher U. 2009. An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase λ. Proc. Natl. Acad. Sci. U. S. A. 106, 18201–18206.
Cadet J., Davies K.J.A., Medeiros M.H.G., Di Mascio P., Wagner J.R. 2017. Formation and repair of oxidatively generated damage in cellular DNA. Free Radic. Biol. Med. 107, 13–34.
Fleming A.M., Burrows C.J. 2017. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic. Biol. Med. 107, 35–52.
Hailer M.K., Slade P.G., Martin B.D., Sugden K.D. 2005. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem. Res. Toxicol. 18, 1378–1383.
Mangerich A., Knutson C.G., Parry N.M., Muthupa-lani S., Ye W., Prestwich E., Cui L., McFaline J.L., Mobley M., Ge Z., Taghizadeh K., Wishnok J.S., Wogan G.N., Fox J.G., Tannenbaum S.R., Dedon P.C. 2012. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. U. S. A. 109, E1820–E1829.
Hu J., de Souza-Pinto N.C., Haraguchi K., Hogue B.A., Jaruga P., Greenberg M.M., Dizdaroglu M., Bohr V.A. 2005. Repair of formamidopyrimidines in DNA involves different glycosylases: Role of the OGG1, NTH1, and NEIL1 enzymes. J. Biol. Chem. 280, 40544–40551.
Dizdaroglu M. 2012. Oxidatively induced DNA damage: Mechanisms, repair and disease. Cancer Lett. 327, 26–47.
Wiederholt C.J., Greenberg M.M. 2002. Fapy ⋅ dG instructs Klenow exo– to misincorporate deoxyadenosine. J. Am. Chem. Soc. 124, 7278–7279.
Kalam M.A., Haraguchi K., Chandani S., Loechler E.L., Moriya M., Greenberg M.M., Basu A.K. 2006. Genetic effects of oxidative DNA damages: Comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 34, 2305–2315.
Pande P., Haraguchi K., Jiang Y.-L., Greenberg M.M., Basu A.K. 2015. Unlike catalyzing error-free bypass of 8-oxodGuo, DNA polymerase λ is responsible for a significant part of Fapy ⋅ dG-induced G→T mutations in human cells. Biochemistry. 54, 1859–1862.
Patro J.N., Wiederholt C.J., Jiang Y.L., Delaney J.C., Essigmann J.M., Greenberg M.M. 2007. Studies on the replication of the ring opened formamidopyrimidine, Fapy ⋅ dG in Escherichia coli. Biochemistry. 46, 10202–10212.
Weledji Y.N., Wiederholt C.J., Delaney M.O., Greenberg M.M. 2008. DNA polymerase bypass in vitro and in E. coli of a C-nucleotide analogue of Fapy ⋅ dG. Bioorg. Med. Chem. 16, 4029–4034.
Imoto S., Patro J.N., Jiang Y.L., Oka N., Greenberg M.M. 2006. Synthesis, DNA polymerase incorporation, and enzymatic phosphate hydrolysis of formamidopyrimidine nucleoside triphosphates. J. Am. Chem. Soc. 128, 14606–14611.
Boiteux S., Gajewski E., Laval J., Dizdaroglu M. 1992. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): Excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 31, 106–110.
Wiederholt C.J., Delaney M.O., Pope M.A., David S.S., Greenberg M.M. 2003. Repair of DNA containing Fapy ⋅ dG and its β-C-nucleoside analogue by formamidopyrimidine DNA glycosylase and MutY. Biochemistry. 42, 9755–9760.
Duarte V., Muller J.G., Burrows C.J. 1999. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 27, 496–502.
Kornyushyna O., Berges A.M., Muller J.G., Burrows C.J. 2002. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment). Biochemistry. 41, 15304–15314.
Zhu J., Fleming A.M., Orendt A.M., Burrows C.J. 2016. pH-dependent equilibrium between 5-guanidinohydantoin and iminoallantoin affects nucleotide insertion opposite the DNA lesion. J. Org. Chem. 81, 351–359.
Beckman J., Wang M., Blaha G., Wang J., Konigsberg W.H. 2010. Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP and dGMP to be inserted opposite guanidinohydantoin. Biochemistry. 49, 8554–8563.
Henderson P.T., Delaney J.C., Muller J.G., Neeley W.L., Tannenbaum S.R., Burrows C.J., Essigmann J.M. 2003. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 42, 9257–9262.
Delaney S., Delaney J.C., Essigmann J.M. 2007. Chemical-biological fingerprinting: Probing the properties of DNA lesions formed by peroxynitrite. Chem. Res. Toxicol. 20, 1718–1729.
Neeley W.L., Delaney S., Alekseyev Y.O., Jarosz D.F., Delaney J.C., Walker G.C., Essigmann J.M. 2007. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 282, 12741–12748.
Huang J., Yennie C.J., Delaney S. 2015. Klenow fragment discriminates against the incorporation of the hyperoxidized dGTP lesion spiroiminodihydantoin into DNA. Chem. Res. Toxicol. 28, 2325–2333.
Leipold M.D., Muller J.G., Burrows C.J., David S.S. 2000. Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme, FPG. Biochemistry. 39, 14984–14992.
Hazra T.K., Muller J.G., Manuel R.C., Burrows C.J., Lloyd R.S., Mitra S. 2001. Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 29, 1967–1974.
Krishnamurthy N., Muller J.G., Burrows C.J., David S.S. 2007. Unusual structural features of hydantoin lesions translate into efficient recognition by Escherichia coli Fpg. Biochemistry. 46, 9355–9365.
Delaney S., Neeley W.L., Delaney J.C., Essigmann J.M. 2007. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 46, 1448–1455.
Leipold M.D., Workman H., Muller J.G., Burrows C.J., David S.S. 2003. Recognition and removal of oxidized guanines in duplex DNA by the base excision repair enzymes hOGG1, yOGG1, and yOGG2. Biochemistry. 42, 11373–11381.
Melamede R.J., Hatahet Z., Kow Y.W., Ide H., Wallace S.S. 1994. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 33, 1255–1264.
Jiang D., Hatahet Z., Melamede R.J., Kow Y.W., Wallace S.S. 1997. Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 272, 32230–32239.
Dizdaroglu M., Burgess S.M., Jaruga P., Hazra T.K., Rodriguez H., Lloyd R.S. 2001. Substrate specificity and excision kinetics of Escherichia coli endonuclease VIII (Nei) for modified bases in DNA damaged by free radicals. Biochemistry. 40, 12150–12156.
Kropachev K.Y., Zharkov D.O., Grollman A.P. 2006. Catalytic mechanism of Escherichia coli endonuclease VIII: Roles of the intercalation loop and the zinc finger. Biochemistry. 45, 12039–12049.
Blaisdell J.O., Hatahet Z., Wallace S.S. 1999. A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G→T transversions. J. Bacteriol. 181, 6396–6402.
Hazra T.K., Izumi T., Venkataraman R., Kow Y.W., Dizdaroglu M., Mitra S. 2000. Characterization of a novel 8-oxoguanine-DNA glycosylase activity in Escherichia coli and identification of the enzyme as endonuclease VIII. J. Biol. Chem. 275, 27762–27767.
Hazra T.K., Izumi T., Boldogh I., Imhoff B., Kow Y.W., Jaruga P., Dizdaroglu M., Mitra S. 2002. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U. S. A. 99, 3523–3528.
Hazra T.K., Kow Y.W., Hatahet Z., Imhoff B., Boldogh I., Mokkapati S.K., Mitra S., Izumi T. 2002. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 277, 30417–30420.
Bandaru V., Sunkara S., Wallace S.S., Bond J.P. 2002. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst.). 1, 517–529.
Morland I., Rolseth V., Luna L., Rognes T., Bjørås M., Seeberg E. 2002. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: An alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 30, 4926–4936.
Takao M., Kanno S.-I., Kobayashi K., Zhang Q.-M., Yonei S., van der Horst G.T.J., Yasui A. 2002. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J. Biol. Chem. 277, 42205–42213.
Rosenquist T.A., Zaika E., Fernandes A.S., Zharkov D.O., Miller H., Grollman A.P. 2003. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst.). 2, 581–591.
Jaruga P., Birincioglu M., Rosenquist T.A., Dizdaroglu M. 2004. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 43, 15909–15914.
Roy L.M., Jaruga P., Wood T.G., McCullough A.K., Dizdaroglu M., Lloyd R.S. 2007. Human polymorphic variants of the NEIL1 DNA glycosylase. J. Biol. Chem. 282, 15790–15798.
Muftuoglu M., de Souza-Pinto N.C., Dogan A., Aamann M., Stevnsner T., Rybanska I., Kirkali G., Dizdaroglu M., Bohr V.A. 2009. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J. Biol. Chem. 284, 9270–9279.
Zou X., Owusu M., Harris R., Jackson S.P., Loizou J.I., Nik-Zainal S. 2018. Validating the concept of mutational signatures with isogenic cell models. Nat. Commun. 9, 1744.
Chan M.K., Ocampo-Hafalla M.T., Vartanian V., Jaruga P., Kirkali G., Koenig K.L., Browng S., Lloyd R.S., Dizdaroglu M., Teebor G.W. 2009. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst.). 8, 786–794.
Hailer M.K., Slade P.G., Martin B.D., Rosenquist T.A., Sugden K.D. 2005. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst.). 4, 41–50.
Krishnamurthy N., Zhao X., Burrows C.J., David S.S. 2008. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 47, 7137–7146.
Liu M., Bandaru V., Bond J.P., Jaruga P., Zhao X., Christov P.P., Burrows C.J., Rizzo C.J., Dizdaroglu M., Wallace S.S. 2010. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Natl Acad. Sci. U. S. A. 107, 4925–4930.
Yeo J., Goodman R.A., Schirle N.T., David S.S., Beal P.A. 2010. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc. Natl. Acad. Sci. U. S. A. 107, 20715–20719.
Zhao X., Krishnamurthy N., Burrows C.J., David S.S. 2010. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 49, 1658–1666.
Redrejo-Rodríguez M., Saint-Pierre C., Couve S., Mazouzi A., Ishchenko A.A., Gasparutto D., Saparbaev M. 2011. New insights in the removal of the hydantoins, oxidation product of pyrimidines, via the base excision and nucleotide incision repair pathways. PLoS One. 6, e21039.
Liu M., Bandaru V., Holmes A., Averill A.M., Cannan W., Wallace S.S. 2012. Expression and purification of active mouse and human NEIL3 proteins. Protein Expr. Purif. 84, 130–139.
Krokeide S.Z., Laerdahl J.K., Salah M., Luna L., Cederkvist F.H., Fleming A.M., Burrows C.J., Dalhus B., Bjørås M. 2013. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst.). 12, 1159–1164.
McKibbin P.L., Fleming A.M., Towheed M.A., van Houten B., Burrows C.J., David S.S. 2013. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J. Am. Chem. Soc. 135, 13851–13861.
Ohtsubo T., Matsuda O., Iba K., Terashima I., Sekiguchi M., Nakabeppu Y. 1998. Molecular cloning of AtMMH, an Arabidopsis thaliana ortholog of the Escherichia coli mutM gene, and analysis of functional domains of its product. Mol. Gen. Genet. 259, 577–590.
Dany A.L., Tissier A. 2001. A functional OGG1 homologue from Arabidopsis thaliana. Mol. Genet. Genomics. 265, 293–301.
García-Ortiz M.-V., Ariza R.R., Roldán-Arjona T. 2001. An OGG1 orthologue encoding a functional 8-oxoguanine DNA glycosylase/lyase in Arabidopsis thaliana. Plant Mol. Biol. 47, 795–804.
Morales-Ruiz T., Birincioglu M., Jaruga P., Rodriguez H., Roldan-Arjona T., Dizdaroglu M. 2003. Arabidopsis thaliana Ogg1 protein excises 8-hydroxyguanine and 2,6-diamino-4-hydroxy-5-formamidopyrimidine from oxidatively damaged DNA containing multiple lesions. Biochemistry. 42, 3089–3095.
Córdoba-Cañero D., Roldán-Arjona T., Ariza R.R. 2014. Arabidopsis ZDP DNA 3′-phosphatase and ARP endonuclease function in 8-oxoG repair initiated by FPG and OGG1 DNA glycosylases. Plant J. 79, 824–834.
Murphy T.M., Guo Y.-Y. 2010. Antimutagenic specificities of two plant glycosylases, oxoguanine glycosylase and formamidopyrimidine glycosylase, assayed in vivo. Biochem. Biophys. Res. Commun. 392, 335–339.
Murphy T.M., George A. 2005. A comparison of two DNA base excision repair glycosylases from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 329, 869–872.
Kathe S.D., Barrantes-Reynolds R., Jaruga P., Newton M.R., Burrows C.J., Bandaru V., Dizdaroglu M., Bond J.P., Wallace S.S. 2009. Plant and fungal Fpg homologs are formamidopyrimidine DNA glycosylases but not 8-oxoguanine DNA glycosylases. DNA Repair (Amst.). 8, 643–653.
Duclos S., Aller P., Jaruga P., Dizdaroglu M., Wallace S.S., Doublié S. 2012. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair (Amst.). 11, 714–725.
Yoshimura K., Ogawa T., Ueda Y., Shigeoka S. 2007. AtNUDX1, an 8-oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphate pyrophosphohydrolase, is responsible for eliminating oxidized nucleotides in Arabidopsis. Plant Cell Physiol. 48, 1438–1449.
Duwat P., de Oliveira R., Ehrlich S.D., Boiteux S. 1995. Repair of oxidative DNA damage in Gram-positive bacteria: The Lactococcus lactis Fpg protein. Microbiology. 141, 411–417.
Castaing B., Fourrey J.-L., Hervouet N., Thomas M., Boiteux S., Zelwer C. 1999. AP site structural determinants for Fpg specific recognition. Nucleic Acids Res. 27, 608–615.
Serre L., Pereira de Jésus K., Boiteux S., Zelwer C., Castaing B. 2002. Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 21, 2854–2865.
Pereira de Jésus K., Serre L., Zelwer C., Castaing B. 2005. Structural insights into abasic site for Fpg specific binding and catalysis: Comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 33, 5936–5944.
Suzuki M., Matsui K., Yamada M., Kasai H., Sofuni T., Nohmi T. 1997. Construction of mutants of Salmonella typhimurium deficient in 8-hydroxyguanine DNA glycosylase and their sensitivities to oxidative mutagens and nitro compounds. Mutat. Res. 393, 233–246.
Mikawa T., Kato R., Sugahara M., Kuramitsu S. 1998. Thermostable repair enzyme for oxidative DNA damage from extremely thermophilic bacterium, Thermus thermophilus HB8. Nucleic Acids Res. 26, 903–910.
Sugahara M., Mikawa T., Kumasaka T., Yamamoto M., Kato R., Fukuyama K., Inoue Y., Kuramitsu S. 2000. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 19, 3857–3869.
Sentürker S., Bauche C., Laval J., Dizdaroglu M. 1999. Substrate specificity of Deinococcus radiodurans Fpg protein. Biochemistry. 38, 9435–9439.
Fromme J.C., Verdine G.L. 2002. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat. Struct. Biol. 9, 544–552.
Qi Y., Spong M.C., Nam K., Banerjee A., Jiralerspong S., Karplus M., Verdine G.L. 2009. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 462, 762–766.
Sung R.-J., Zhang M., Qi Y., Verdine G.L. 2012. Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM. J. Biol. Chem. 287, 18044–18054.
Sung R.-J., Zhang M., Qi Y., Verdine G.L. 2013. Structural and biochemical analysis of DNA helix invasion by the bacterial 8-oxoguanine DNA glycosylase MutM. J. Biol. Chem. 288, 10012–10023.
Nagorska K., Silhan J., Li Y., Pelicic V., Freemont P.S., Baldwin G.S., Tang C.M. 2012. A network of enzymes involved in repair of oxidative DNA damage in Neisseria meningitidis. Mol. Microbiol. 83, 1064–1079.
Souza Arantes L., Gonçalves Vila Nova L., Resende B.C., Bitar M., Vale Coelho I.E., Miyoshi A., Azevedo V.A., dos Santos L.L., Machado C.R., de Oliveira Lopes D. 2016. The Corynebacterium pseudotuberculosis genome contains two formamidopyrimidine-DNA glycosylase enzymes, only one of which recognizes and excises 8-oxoguanine lesion. Gene. 575, 233–243.
Davidsen T., Bjørås M., Seeberg E.C., Tønjum T. 2005. Antimutator role of DNA glycosylase MutY in pathogenic Neisseria species. J. Bacteriol. 187, 2801–2809.
Eutsey R., Wang G., Maier R.J. 2007. Role of a MutY DNA glycosylase in combating oxidative DNA damage in Helicobacter pylori. DNA Repair (Amst.). 6, 19–26.
Robles A.G., Reid K., Roy F., Fletcher H.M. 2011. Porphyromonas gingivalis mutY is involved in the repair of oxidative stress-induced DNA mispairing. Mol. Oral Microbiol. 26, 175–186.
Eberle R.J., Coronado M.A., Caruso I.P., Lopes D.O., Miyoshi A., Azevedo V., Arni R.K. 2015. Chemical and thermal influence of the [4Fe–4S]2+ cluster of A/G-specific adenine glycosylase from Corynebacterium pseudotuberculosis. Biochim. Biophys. Acta. 1850, 393–400.
Cançado de Faria R., Gonçalves Vila-Nova L., Bitar M., Carvalho Resende B., Sousa Arantes L., Basso Rebelato A., Carvalho Azevedo V.A., Franco G.R., Machado C.R., dos Santos L.L., de Oliveira Lopes D. 2016. Adenine glycosylase MutY of Corynebacterium pseudotuberculosis presents the antimutator phenotype and evidences of glycosylase/AP lyase activity in vitro. Infect. Genet. Evol. 44, 318–329.
Dos Vultos T., Blázquez J., Rauzier J., Matic I., Gicquel B. 2006. Identification of Nudix hydrolase family members with an antimutator role in Mycobacterium tuberculosis and Mycobacterium smegmatis. J. Bacteriol. 188, 3159–3161.
Patil A.G.G., Sang P.B., Govindan A., Varshney U. 2013. Mycobacterium tuberculosis MutT1 (Rv2985) and ADPRase (Rv1700) proteins constitute a two-stage mechanism of 8-oxo-dGTP and 8-oxo-GTP detoxification and adenosine to cytidine mutation avoidance. J. Biol. Chem. 288, 11252–11262.
Arif S.M., Patil A.G., Varshney U., Vijayan M. 2017. Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association. Acta Crystallogr. D Struct. Biol. 73, 349–364.
Sasaki M., Yonemura Y., Kurusu Y. 2000. Genetic analysis of Bacillus subtilis mutator genes. J. Gen. Appl. Microbiol. 46, 183–187.
Castellanos-Juárez F.X., Álvarez-Álvarez C., Yasbin R.E., Setlow B., Setlow P., Pedraza-Reyes M. 2006. YtkD and MutT protect vegetative cells but not spores of Bacillus subtilis from oxidative stress. J. Bacteriol. 188, 2285–2289.
Vidales L.E., Cárdenas L.C., Robleto E., Yasbin R.E., Pedraza-Reyes M. 2009. Defects in the error prevention oxidized guanine system potentiate stationary-phase mutagenesis in Bacillus subtilis. J. Bacteriol. 191, 506–513.
Lian K., Leiros H.-K.S., Moe E. 2015. MutT from the fish pathogen Aliivibrio salmonicida is a cold-active nucleotide-pool sanitization enzyme with unexpectedly high thermostability. FEBS Open Bio. 5, 107–116.
Santos-Escobar F., Gutiérrez-Corona J.F., Pedraza-Reyes M. 2014. Role of Bacillus subtilis error prevention oxidized guanine system in counteracting hexavalent chromium-promoted oxidative DNA damage. Appl. Environ. Microbiol. 80, 5493–5502.
Oliver A., Sánchez J.M., Blázquez J. 2002. Characterization of the GO system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 217, 31–35.
Sanders L.H., Sudhakaran J., Sutton M.D. 2009. The GO system prevents ROS-induced mutagenesis and killing in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 294, 89–96.
Davidsen T., Amundsen E.K., Rødland E.A., Tønjum T. 2007. DNA repair profiles of disease-associated isolates of Neisseria meningitidis. FEMS Immunol. Med. Microbiol. 49, 243–251.
Davidsen T., Tuven H.K., Bjørås M., Rødland E.A., Tønjum T. 2007. Genetic interactions of DNA repair pathways in the pathogen Neisseria meningitidis. J. Bacteriol. 189, 5728–5737.
Canfield G.S., Schwingel J.M., Foley M.H., Vore K.L., Boonanantanasarn K., Gill A.L., Sutton M.D., Gill S.R. 2013. Evolution in fast forward: A potential role for mutators in accelerating Staphylococcus aureus pathoadaptation. J. Bacteriol. 195, 615–628.
Martins-Pinheiro M., Oliveira A.R., Valencia A.O., Fernandez-Silva F.S., Silva L.G., Lopes-Kulishev C.O., Italiani V.C.S., Marques M.V., Menck C.F., Galhardo R.S. 2017. Molecular characterization of Caulobacter crescentus mutator strains. Gene. 626, 251–257.
Fernández-Silva F.S., Schulz M.L., Alves I.R., Freitas R.R., Paes da Rocha R., Lopes-Kulishev C.O., Medeiros M.H.G., Galhardo R.S. 2020. Contribution of GO system glycosylases to mutation prevention in Caulobacter crescentus. Environ. Mol. Mutagen. 61, 246–255.
Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 393, 537–544.
Jain R., Kumar P., Varshney U. 2007. A distinct role of formamidopyrimidine DNA glycosylase (MutM) in down-regulation of accumulation of G, C mutations and protection against oxidative stress in mycobacteria. DNA Repair (Amst.). 6, 1774–1785.
Sidorenko V.S., Rot M.A., Filipenko M.L., Nevinsky G.A., Zharkov D.O. 2008. Novel DNA glycosylases from Mycobacterium tuberculosis. Biochemistry (Moscow). 73 (4), 442–450.
Kurthkoti K., Srinath T., Kumar P., Malshetty V.S., Sang P.B., Jain R., Manjunath R., Varshney U. 2010. A distinct physiological role of MutY in mutation prevention in mycobacteria. Microbiology. 156, 88–93.
Kurthkoti K., Varshney U. 2011. Base excision and nucleotide excision repair pathways in mycobacteria. Tuberculosis. 91, 533–543.
Kurthkoti K., Varshney U. 2012. Distinct mechanisms of DNA repair in mycobacteria and their implications in attenuation of the pathogen growth. Mech. Ageing Dev. 133, 138–146.
Hassim F., Papadopoulos A.O., Kana B.D., Gordhan B.G. 2015. A combinatorial role for MutY and Fpg DNA glycosylases in mutation avoidance in Mycobacterium smegmatis. Mutat. Res. 779, 24–32.
Kurthkoti K., Kumar P., Jain R., Varshney U. 2008. Important role of the nucleotide excision repair pathway in Mycobacterium smegmatis in conferring protection against commonly encountered DNA-damaging agents. Microbiology. 154, 2776–2785.
Mestre O., Hurtado-Ortiz R., Dos Vultos T., Namouchi A., Cimino M., Pimentel M., Neyrolles O., Gicquel B. 2013. High throughput phenotypic selection of Mycobacterium tuberculosis mutants with impaired resistance to reactive oxygen species identifies genes important for intracellular growth. PLoS One. 8, e53486.
Olsen I., Balasingham S.V., Davidsen T., Debebe E., Rødland E.A., van Soolingen D., Kremer K., Alseth I., Tønjum T. 2009. Characterization of the major formamidopyrimidine-DNA glycosylase homolog in Mycobacterium tuberculosis and its linkage to variable tandem repeats. FEMS Immunol. Med. Microbiol. 56, 151–161.
Guo Y., Bandaru V., Jaruga P., Zhao X., Burrows C.J., Iwai S., Dizdaroglu M., Bond J.P., Wallace S.S. 2010. The oxidative DNA glycosylases of Mycobacterium tuberculosis exhibit different substrate preferences from their Escherichia coli counterparts. DNA Repair (Amst.). 9, 177–190.
Kinoshita-Daitoku R., Kiga K., Sanada T., Ogura Y., Bo Z., Iida T., Yokomori R., Kuroda E., Tanaka M., Sood A., Suzuki T., Nakai K., Hayashi T., Mimuro H. 2020. Mutational diversity in mutY-deficient Helicobacter pylori and its effect on adaptation to the gastric environment. Biochem. Biophys. Res. Commun. 525, 806–811.
Eskra L., Canavessi A., Carey M., Splitter G. 2001. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect. Immun. 69, 7736–7742.
Baharoglu Z., Krin E., Mazel D. 2013. RpoS plays a central role in the SOS induction by sub-lethal aminoglycoside concentrations in Vibrio cholerae. PLoS Genet. 9, e1003421.
Dupuy P., Howlader M., Glickman M.S. 2020. A multilayered repair system protects the mycobacterial chromosome from endogenous and antibiotic-induced oxidative damage. Proc. Natl. Acad. Sci. U. S. A. 117, 19517–19527.
Oliver A., Cantón R., Campo P., Baquero F., Blázquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 288, 1251–1253.
Mandsberg L.F., Ciofu O., Kirkby N., Christiansen L.E., Poulsen H.E., Høiby N. 2009. Antibiotic resistance in Pseudomonas aeruginosa strains with increased mutation frequency due to inactivation of the DNA oxidative repair system. Antimicrob. Agents Chemother. 53, 2483–2491.
Morero N.R., Argaraña C.E. 2009. Pseudomonas aeruginosa deficient in 8-oxodeoxyguanine repair system shows a high frequency of resistance to ciprofloxacin. FEMS Microbiol. Lett. 290, 217–226.
Mandsberg L.F., Maciá M.D., Bergmann K.R., Christiansen L.E., Alhede M., Kirkby N., Høiby N., Oliver A., Ciofu O. 2011. Development of antibiotic resistance and up-regulation of the antimutator gene pfpI in mutator Pseudomonas aeruginosa due to inactivation of two DNA oxidative repair genes (mutY, mutM). FEMS Microbiol. Lett. 324, 28–37.
Couce A., Alonso-Rodriguez N., Costas C., Oliver A., Blázquez J. 2016. Intrapopulation variability in mutator prevalence among urinary tract infection isolates of Escherichia coli. Clin. Microbiol. Infect. 22, 566.
Perrin A., Larsonneur E., Nicholson A.C., Edwards D.J., Gundlach K.M., Whitney A.M., Gulvik C.A., Bell M.E., Rendueles O., Cury J., Hugon P., Clermont D., Enouf V., Loparev V., Juieng P., et al. 2017. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun. 8, 15483.
Bridges B.A. 1995. mutY ‘directs’ mutation? Nature. 375, 741.
Bridges B.A., Timms A.R. 1997. Mutation in Escherichia coli under starvation conditions: A new pathway leading to small deletions in strains defective in mismatch correction. EMBO J. 16, 3349–3356.
Notley-McRobb L., Pinto R., Seeto S., Ferenci T. 2002. Regulation of mutY and nature of mutator mutations in Escherichia coli populations under nutrient limitation. J. Bacteriol. 184, 739–745.
Raynes Y., Sniegowski P.D. 2014. Experimental evolution and the dynamics of genomic mutation rate modifiers. Heredity. 113, 375–380.
Couce A., Tenaillon O. 2019. Mutation bias and GC content shape antimutator invasions. Nat. Commun. 10, 3114.
Lu A.-L., Fawcett W.P. 1998. Characterization of the recombinant MutY homolog, an adenine DNA glycosylase, from yeast Schizosaccharomyces pombe. J. Biol. Chem. 273, 25098–25105.
Chang D.-Y., Lu A.-L. 2002. Functional interaction of MutY homolog with proliferating cell nuclear antigen in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 277, 11853–11858.
Chang D.-Y., Lu A.-L. 2005. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 280, 408–417.
Girard P.-M., Guibourt N., Boiteux S. 1997. The Ogg1 protein of Saccharomyces cerevisiae: A 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res. 25, 3204–3211.
Karahalil B., Girard P.-M., Boiteux S., Dizdaroglu M. 1998. Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation- or hydrogen peroxide/metal ion-generated free radicals. Nucleic Acids Res. 26, 1228–1232.
Thomas D., Scot A.D., Barbey R., Padula M., Boiteux S. 1997. Inactivation of OGG1 increases the incidence of G ⋅ C→T ⋅ A transversions in Saccharomyces cerevisiae: Evidence for endogenous oxidative damage in DNA in eukaryotic cells. Mol. Gen. Genet. 254, 171–178.
Guibourt N., Boiteux S. 2000. Expression of the Fpg protein of Escherichia coli in Saccharomyces cerevisiae: Effects on spontaneous mutagenesis and sensitivity to oxidative DNA damage. Biochimie. 82, 59–64.
Scott A.D., Neishabury M., Jones D.H., Reed S.H., Boiteux S., Waters R. 1999. Spontaneous mutation, oxidative DNA damage, and the roles of base and nucleotide excision repair in the yeast Saccharomyces cerevisiae. Yeast. 15, 205–218.
Ni T.T., Marsischky G.T., Kolodner R.D. 1999. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol. Cell. 4, 439–444.
Shockley A.H., Doo D.W., Rodriguez G.P., Crouse G.F. 2013. Oxidative damage and mutagenesis in Saccharomyces cerevisiae: Genetic studies of pathways affecting replication fidelity of 8-oxoguanine. Genetics. 195, 359–367.
Earley M.C., Crouse G.F. 1998. The role of mismatch repair in the prevention of base pair mutations in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 95, 15487–15491.
Pavlov Y.I., Mian I.M., Kunkel T.A. 2003. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr. Biol. 13, 744–748.
Haracska L., Yu S.-L., Johnson R.E., Prakash L., Prakash S. 2000. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 25, 458–461.
de Padula M., Slezak G., Auffret van Der Kemp P., Boiteux S. 2004. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res. 32, 5003–5010.
Yung C.-W., Okugawa Y., Otsuka C., Okamoto K., Arimoto S., Loakes D., Negishi K., Negishi T. 2008. Influence of neighbouring base sequences on the mutagenesis induced by 7,8-dihydro-8-oxoguanine in yeast. Mutagenesis. 23, 509–513.
Auffret van der Kemp P., de Padula M., Burguiere-Slezak G., Ulrich H.D., Boiteux S. 2009. PCNA monoubiquitylation and DNA polymerase η ubiquitin-binding domain are required to prevent 8-oxoguanine-induced mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 37, 2549–2559.
Kaniak A., Dzierzbicki P., Rogowska A.T., Malc E., Fikus M., Ciesla Z. 2009. Msh1p counteracts oxidative lesion-induced instability of mtDNA and stimulates mitochondrial recombination in Saccharomyces cerevisiae. DNA Repair (Amst.). 8, 318–329.
Bruner S.D., Nash H.M., Lane W.S., Verdine G.L. 1998. Repair of oxidatively damaged guanine in Saccharomyces cerevisiae by an alternative pathway. Curr. Biol. 8, 393–403.
Sentürker S., Dizdaroglu M., Auffret van der Kemp P., You H.J., Doetsch P.W., Boiteux S. 1998. Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 26, 5270–5276.
Eide L., Bjørås M., Pirovano M., Alseth I., Berdal K.G., Seeberg E. 1996. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93, 10735–10740.
You H.J., Swanson R.L., Doetsch P.W. 1998. Saccharomyces cerevisiae possesses two functional homologues of Escherichia coli endonuclease III. Biochemistry. 37, 6033–6040.
You H.J., Swanson R.L., Harrington C., Corbett A.H., Jinks-Robertson S., Sentürker S., Wallace S.S., Boiteux S., Dizdaroglu M., Doetsch P.W. 1999. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 38, 11298–11306.
Ishchenko A.A., Yang X., Ramotar D., Saparbaev M. 2005. The 3′ → 5′ exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 6380–6390.
Mazurek A., Berardini M., Fishel R. 2002. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J. Biol. Chem. 277, 8260–8266.
Larson E.D., Iams K., Drummond J.T. 2003. Strand-specific processing of 8-oxoguanine by the human mismatch repair pathway: inefficient removal of 8-oxoguanine paired with adenine or cytosine. DNA Repair (Amst.). 2, 1199–1210.
Macpherson P., Barone F., Maga G., Mazzei F., Karran P., Bignami M. 2005. 8-Oxoguanine incorporation into DNA repeats in vitro and mismatch recognition by MutSα. Nucleic Acids Res. 33, 5094–5105.
Pitsikas P., Lee D., Rainbow A.J. 2007. Reduced host cell reactivation of oxidative DNA damage in human cells deficient in the mismatch repair gene hMSH2. Mutagenesis. 22, 235–243.
Viel A., Bruselles A., Meccia E., Fornasarig M., Quaia M., Canzonieri V., Policicchio E., Urso E.D., Agostini M., Genuardi M., Lucci-Cordisco E., Venesio T., Martayan A., Diodoro M.G., Sanchez-Mete L., et al. 2017. A specific mutational signature associated with DNA 8-oxoguanine persistence in MUTYH-defective colorectal cancer. EBioMedicine. 20, 39–49.
Colussi C., Parlanti E., Degan P., Aquilina G., Barnes D., Macpherson P., Karran P., Crescenzi M., Dogliotti E., Bignami M. 2002. The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr. Biol. 12, 912–918.
McDowell H.D., Carney J.P., Wilson T.M. 2008. Inhibition of the 5′ to 3′ exonuclease activity of hEXO1 by 8-oxoguanine. Environ. Mol. Mutagen. 49, 388–398.
Gu Y., Parker A., Wilson T.M., Bai H., Chang D.-Y., Lu A.-L. 2002. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 277, 11135–11142.
Repmann S., Olivera-Harris M., Jiricny J. 2015. Influence of oxidized purine processing on strand directionality of mismatch repair. J. Biol. Chem. 290, 9986–9999.
Gogos A., Clarke N.D. 1999. Characterization of an 8-oxoguanine DNA glycosylase from Methanococcus jannaschii. J. Biol. Chem. 274, 30447–30450.
Faucher F., Duclos S., Bandaru V., Wallace S.S., Doublié S. 2009. Crystal structures of two archaeal 8‑oxoguanine DNA glycosylases provide structural insight into guanine/8-oxoguanine distinction. Structure. 17, 703–712.
Faucher F., Wallace S.S., Doublié S. 2010. The C-terminal lysine of Ogg2 DNA glycosylases is a major molecular determinant for guanine/8-oxoguanine distinction. J. Mol. Biol. 397, 46–56.
Chung J.H., Suh M.-J., Park Y.I., Tainer J.A., Han Y.S. 2001. Repair activities of 8-oxoguanine DNA glycosylase from Archaeoglobus fulgidus, a hyperthermophilic archaeon. Mutat. Res. 486, 99–111.
Fujii M., Hata C., Ukita M., Fukushima C., Matsuura C., Kawashima-Ohya Y., Tomobe K., Kawashima T. 2016. Characterization of a thermostable 8-oxoguanine DNA glycosylase specific for GO/N mismatches from the thermoacidophilic archaeon Thermoplasma volcanium. Archaea. 2016, 8734894.
Sartori A.A., Lingaraju G.M., Hunziker P., Winkler F.K., Jiricny J. 2004. Pa-AGOG, the founding member of a new family of archaeal 8-oxoguanine DNA-glycosylases. Nucleic Acids Res. 32, 6531–6539.
Lingaraju G.M., Sartori A.A., Kostrewa D., Prota A.E., Jiricny J., Winkler F.K. 2005. A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure. Structure. 13, 87–98.
Lingaraju G.M., Prota A.E., Winkler F.K. 2009. Mutational studies of Pa-AGOG DNA glycosylase from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. DNA Repair (Amst.). 8, 857–864.
Zhang L., Li Y., Shi H., Zhang D., Yang Z., Oger P., Zheng J. 2019. Biochemical characterization and mutational studies of the 8-oxoguanine DNA glycosylase from the hyperthermophilic and radioresistant archaeon Thermococcus gammatolerans. Appl. Microbiol. Biotechnol. 103, 8021–8033.
Gehring A.M., Zatopek K.M., Burkhart B.W., Potapov V., Santangelo T.J., Gardner A.F. 2020. Biochemical reconstitution and genetic characterization of the major oxidative damage base excision DNA repair pathway in Thermococcus kodakarensis. DNA Repair (Amst.). 86, 102767.
White M.F., Allers T. 2018. DNA repair in the archaea: An emerging picture. FEMS Microbiol. Rev. 42, 514–526.
Ishino S., Nishi Y., Oda S., Uemori T., Sagara T., Takatsu N., Yamagami T., Shirai T., Ishino Y. 2016. Identification of a mismatch-specific endonuclease in hyperthermophilic Archaea. Nucleic Acids Res. 44, 2977–2986.
Al-Tassan N., Chmiel N.H., Maynard J., Fleming N., Livingston A.L., Williams G.T., Hodges A.K., Davies D.R., David S.S., Sampson J.R., Cheadle J.P. 2002. Inherited variants of MYH associated with somatic G:C → T:A mutations in colorectal tumors. Nat. Genet. 30, 227–232.
Chmiel N.H., Livingston A.L., David S.S. 2003. Insight into the functional consequences of inherited variants of the hMYH adenine glycosylase associated with colorectal cancer: Complementation assays with hMYH variants and pre-steady-state kinetics of the corresponding mutated E. coli enzymes. J. Mol. Biol. 327, 431–443.
Livingston A.L., Kundu S., Pozzi M.H., Anderson D.W., David S.S. 2005. Insight into the roles of tyrosine 82 and glycine 253 in the Escherichia coli adenine glycosylase MutY. Biochemistry. 44, 14179–14190.
Pope M.A., Chmiel N.H., David S.S. 2005. Insight into the functional consequences of hMYH variants associated with colorectal cancer: Distinct differences in the adenine glycosylase activity and the response to AP endonucleases of Y150C and G365D murine MYH. DNA Repair (Amst.). 4, 315–325.
Jones S., Emmerson P., Maynard J., Best J.M., Jordan S., Williams G.T., Sampson J.R., Cheadle J.P. 2002. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C→T:A mutations. Hum. Mol. Genet. 11, 2961–2967.
Sieber O.M., Lipton L., Crabtree M., Heinimann K., Fidalgo P., Phillips R.K.S., Bisgaard M.-L., Orntoft T.F., Aaltonen L.A., Hodgson S.V., Thomas H.J.W., Tomlinson I.P.M. 2003. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N. Engl. J. Med. 348, 791–799.
Sampson J.R., Dolwani S., Jones S., Eccles D., Ellis A., Evans D.G., Frayling I., Jordan S., Maher E.R., Mak T., Maynard J., Pigatto F., Shaw J., Cheadle J.P. 2003. Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet. 362, 39–41.
Farrington S.M., Tenesa A., Barnetson R., Wiltshire A., Prendergast J., Porteous M., Campbell H., Dunlop M.G. 2005. Germline susceptibility to colorectal cancer due to base-excision repair gene defects. Am. J. Hum. Genet. 77, 112–119.
Nielsen M., Infante E., Brand R. 2019. in GeneReviews™ [Internet]. Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., Eds. Seattle, WA: Univ. of Washington Seattle.
Short E., Sampson J. 2019. The role of inherited genetic variants in colorectal polyposis syndromes. Adv. Genet. 103, 183–217.
Curia M.C., Catalano T., Aceto G.M. 2020. MUTYH: Not just polyposis. World J. Clin. Oncol. 11, 428–449.
Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.-L., Boyault S., Burkhardt B., Butler A.P., Caldas C., Davies H.R., et al. 2013. Signatures of mutational processes in human cancer. Nature. 500, 415–421.
Simonelli V., Camerini S., Mazzei F., Van Loon B., Allione A., D’Errico M., Barone F., Minoprio A., Ricceri F., Guarrera S., Russo A., Dalhus B., Crescenzi M., Hübscher U., Bjørås M., et al. 2013. Genotype–phenotype analysis of S326C OGG1 polymorphism: a risk factor for oxidative pathologies. Free Radic. Biol. Med. 63, 401–409.
Kang S.W., Kim S.K., Park H.J., Chung J.-H., Ban J.Y. 2017. Human 8-oxoguanine DNA glycosylase gene polymorphism (Ser326Cys) and cancer risk: Updated meta-analysis. Oncotarget. 8, 44761–44775.
Kohno T., Shinmura K., Tosaka M., Tani M., Kim S.-R., Sugimura H., Nohmi T., Kasai H., Yokota J. 1998. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 16, 3219–3225.
Luna L., Rolseth V., Hildrestrand G.A., Otterlei M., Dantzer F., Bjørås M., Seeberg E. 2005. Dynamic relocalization of hOGG1 during the cell cycle is disrupted in cells harbouring the hOGG1-Cys326 polymorphic variant. Nucleic Acids Res. 33, 1813–1824.
Hill J.W., Evans M.K. 2006. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Res. 34, 1620–1632.
Bravard A., Vacher M., Moritz E., Vaslin L., Hall J., Epe B., Radicella J.P. 2009. Oxidation status of human OGG1-S326C polymorphic variant determines cellular DNA repair capacity. Cancer Res. 69, 3642–3649.
Kershaw R.M., Hodges N.J. 2012. Repair of oxidative DNA damage is delayed in the Ser326Cys polymorphic variant of the base excision repair protein OGG1. Mutagenesis. 27, 501–510.
Moritz E., Pauly K., Bravard A., Hall J., Radicella J.P., Epe B. 2014. hOGG1-Cys326 variant cells are hypersensitive to DNA repair inhibition by nitric oxide. Carcinogenesis. 35, 1426–1433.
Yakushiji H., Maraboeuf F., Takahashi M., Deng Z.-S., Kawabata S.-I., Nakabeppu Y., Sekiguchi M. 1997. Biochemical and physicochemical characterization of normal and variant forms of human MTH1 protein with antimutagenic activity. Mutat. Res. 384, 181–194.
Sakai Y., Oda H., Yoshimura D., Furuichi M., Kang D., Iwai S., Hara T., Nakabeppu Y. 2006. The GT to GC single nucleotide polymorphism at the beginning of an alternative exon 2C of human MTH1 gene confers an amino terminal extension that functions as a mitochondrial targeting signal. J. Mol. Med. 84, 660–670.
Kimura Y., Oda S., Egashira A., Kakeji Y., Baba H., Nakabeppu Y., Maehara Y. 2004. A variant form of hMTH1, a human homologue of the E coli mutT gene, correlates with somatic mutation in the p53 tumour suppressor gene in gastric cancer patients. J. Med. Genet. 41, e57.
Miyako K., Kohno H., Ihara K., Kuromaru R., Matsuura N., Hara T. 2004. Association study of human MTH1 gene polymorphisms with type 1 diabetes mellitus. Endocr. J. 51, 493–498.
Kohno T., Sakiyama T., Kunitoh H., Goto K., Nishiwaki Y., Saito D., Hirose H., Eguchi T., Yanagitani N., Saito R., Sasaki-Matsumura R., Mimaki S., Toyama K., Yamamoto S., Kuchiba A., et al. 2006. Association of polymorphisms in the MTH1 gene with small cell lung carcinoma risk. Carcinogenesis. 27, 2448–2454.
Cao L., Zhou W., Zhu Y., Guo W., Cai Z., He X., Xie Y., Li X., Zhu D., Wang Y. 2013. Combined analysis of polymorphism variants in hMTH1, hOGG1 and MUTYH genes on the risk of type 2 diabetes in the Chinese population. Gene. 519, 50–54.
Fleming A.M., Burrows C.J. 2017. 8-Oxo-7,8-dihydroguanine, friend and foe: Epigenetic-like regulator versus initiator of mutagenesis. DNA Repair (Amst.). 56, 75–83.
Fleming A.M., Ding Y., Burrows C.J. 2017. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 114, 2604–2609.
Tahara Y.-K., Kietrys A.M., Hebenbrock M., Lee Y., Wilson D.L., Kool E.T. 2019. Dual inhibitors of 8-oxo-guanine surveillance by OGG1 and NUDT1. ACS Chem. Biol. 14, 2606–2615.
Warpman Berglund U., Sanjiv K., Gad H., Kalderén C., Koolmeister T., Pham T., Gokturk C., Jafari R., Maddalo G., Seashore-Ludlow B., Chernobrovkin A., Manoilov A., Pateras I.S., Rasti A., Jemth A.-S., et al. 2016. Validation and development of MTH1 inhibitors for treatment of cancer. Ann. Oncol. 27, 2275–2283.
Hua X., Sanjiv K., Gad H., Pham T., Gokturk C., Rasti A., Zhao Z., He K., Feng M., Zang Y., Zhang J., Xia Q., Helleday T., Warpman Berglund U. 2019. Karonudib is a promising anticancer therapy in hepatocellular carcinoma. Ther. Adv. Med. Oncol. 11, 1758835919866960.
Magkouta S.F., Pappas A.G., Vaitsi P.C., Agioutantis P.C., Pateras I.S., Moschos C.A., Iliopoulou M.P., Kosti C.N., Loutrari H.V., Gorgoulis V.G., Kalomenidis I.T. 2020. MTH1 favors mesothelioma progression and mediates paracrine rescue of bystander endothelium from oxidative damage. JCI Insight. 5, e134885.
Samaranayake G.J., Huynh M., Rai P. 2017. MTH1 as a chemotherapeutic target: the elephant in the room. Cancers. 9, 47.
Mechetin G.V., Endutkin A.V., Diatlova E.A., Zharkov D.O. 2020. Inhibitors of DNA glycosylases as prospective drugs. Int. J. Mol. Sci. 21, 3118.
Funding
The work was supported by grant no. 19-74-00068 from the Russian Science Foundation (GO systems in bacteria) and partially by grant no. 17-00-00261/17-00-00265 from the Russian Foundation for Basic Research (GO systems in eukaryotes).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors state that there is no conflict of interest.
This article does not contain any studies with the use of humans as objects of research.
Additional information
Translated by A. Levina
Rights and permissions
About this article
Cite this article
Endutkin, A.V., Zharkov, D.O. GO System, a DNA Repair Pathway to Cope with Oxidative Damage. Mol Biol 55, 193–210 (2021). https://doi.org/10.1134/S0026893321020072
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893321020072