Abstract
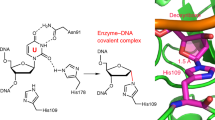
The human N-glycosylases SMUG1 and MBD4 catalyze the removal of uracil residues from DNA resulting from cytosine deamination or replication errors. For polymorphic variants of SMUG1 (G90C, P240H, N244S, N248Y) and the MBD4cat catalytic domain (S470L, G507S, R512W, H557D), the structures of enzyme-substrate complexes were obtained by molecular dynamic simulation. It was experimentally found that the SNP variants of SMUG1, N244S and N248Y, had increased catalytic activity compared to the wild-type enzyme, probably due to the acceleration of the dissociation of the enzyme–product complex and an increase in the enzyme turnover rate. All other SNP variants of SMUG1 (G90C, P240H) and MBD4cat, in which amino acid substitutions disrupted the substrate binding region and/or active site, had significantly lower catalytic activity than the wild-type enzymes.





Similar content being viewed by others
REFERENCES
Lari S.-U., Chen C.-Y., Vertéssy B.G., Morré J., Bennett S.E. 2006. Quantitative determination of uracil residues in Escherichia coli DNA: contribution of ung, dug, and dut genes to uracil avoidance. DNA Repair (Amst.). 5, 1407–1420.
Lewis C.A., Crayle J., Zhou S., Swanstrom R., Wolfenden R.,Wolfenden R. 2016. Cytosine deamination and the precipitous decline of spontaneous mutation during Earth’s history. Proc. Natl. Acad. Sci. U. S. A. 113, 8194–8199.
Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature. 362, 709–715.
Jaszczur M., Bertram J.G., Pham P., Scharff M.D., Goodman M.F. 2013. AID and Apobec3G haphazard deamination and mutational diversity. Cell. Mol. Life Sci. 70, 3089–3108.
Rebhandl S., Hümer M., Greil R., Geisberger R. 2015. AID/APOBEC deaminases and cancer. Oncoscience. 2, 320–333.
Ladner R.D. 2001. The role of dUTPase and uracil-DNA repair in cancer chemotherapy. Curr. Protein Pept. Sci. 2, 361–370.
Krokan H.E., Drabløs F., Slupphaug G. 2002. Uracil in DNA: Occurrence, consequences and repair. Oncogene. 21, 8935–8948.
Jacobs A.L., Schar P. 2012. DNA glycosylases: In DNA repair and beyond. Chromosoma. 121, 1–20.
Visnes T., Doseth B., Pettersen H.S., Hagen L., Sousa M.M., Akbari M., Otterlei M., Kavli B., Slupphaug G., Krokan H.E. 2009. Uracil in DNA and its processing by different DNA glycosylases. Philos. Trans. R. Soc. B. 364, 563–568.
Dawson N.L., Lewis T.E., Das S., Lees J.G., Lee D., Ashford P., Orengo C.A., Sillitoe I. 2017. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 45, D289–D295.
Wibley J.E.A., Waters T.R., Haushalter K., Verdine G.L., Pearl L.H. 2003. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol. Cell. 11, 1647–1659.
Kavli B., Sundheim O., Akbari M., Otterlei M., Nilsen H., Skorpen F., Aas P.A., Hagen L., Krokan H.E., Slupphaug G. 2002. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 277, 39926–39936.
Kavli B., Otterlei M., Slupphaug G., Krokan H.E. 2007. Uracil in DNA: General mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst.). 6, 505–516.
Zhang Z., Shen J., Yang Y., Li J., Cao W., Xie W. 2016. Structural basis of substrate specificity in Geobacter metallireducens SMUG1. ACS Chem. Biol. 11, 1729–1736.
Kuznetsova A.A., Iakovlev D.A., Misovets I.V., Ishchenko A.A., Saparbaev M.K., Kuznetsov N.A., Fedorova O.S. 2017. Pre-steady-state kinetic analysis of damage recognition by human single-strand selective monofunctional uracil-DNA glycosylase SMUG1. Mol. Biosyst. 13, 2638–2649.
Matsubara M., Tanaka T., Terato H., Ohmae E., Izumi S., Katayanagi K., Ide H. 2004. Mutational analysis of the damage-recognition and catalytic mechanism of human SMUG1 DNA glycosylase. Nucleic Acids Res. 32, 5291–5302.
Iakovlev D.A., Alekseeva I. V., Vorobjev Y.N., Kuznetsov N.A., Fedorova O.S. 2019. The role of active-site residues Phe98, HiS239, and Arg243 in DNA binding and in the catalysis of human uracil-DNA glycosylase SMUG1. Molecules. 24 (17), 3133.
Iakovlev D.A., Alekseeva I.V., Kuznetsov N.A., Fedo-rova O.S. 2020. Role of Arg243 and His239 residues in the recognition of damaged nucleotides by human uracil-DNA glycosylase SMUG1. Biochemistry (Moscow). 85 (5), 594‒603.
Sjolund A.B., Senejani A.G., Sweasy J.B. 2013. MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res. 743–744, 12–25.
Turner D.P., Cortellino S., Schupp J.E., Caretti E., Loh T., Kinsella T.J., Bellacosa A. 2006. The DNA N‑glycosylase MED1 exhibits preference for halogenated pyrimidines and is involved in the cytotoxicity of 5-iododeoxyuridine. Cancer Res. 66 (15), 7686–7693.
Morera S., Grin I., Vigouroux A., Couve S., Henriot V., Saparbaev M., Ishchenko A.A. 2012. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res. 40, 9917–9926.
Hill P.W., Amouroux R., Hajkova P. 2014. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story. Genomics. 104, 324–333.
Rai K., Huggins I.J., James S.R., Karpf A.R., Jones D.A., Cairns B.R. 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell. 135, 1201–1212.
Walavalkar N.M., Cramer J.M., Buchwald W.A., Scarsdale J.N., Williams D.C. 2014. Solution structure and intramolecular exchange of methyl-cytosine binding domain protein 4 (MBD4) on DNA suggests a mechanism to scan for mCpG/TpG mismatches. Nucleic Acids Res. 42, 11218–11232.
Manvilla B.A., Maiti A., Begley M.C., Toth E.A., Drohat A.C. 2012. Crystal structure of human methyl-binding domain IV glycosylase bound to abasic DNA. J. Mol. Biol. 420, 164–175.
Hashimoto H., Zhang X., Cheng X. 2012. Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation. Nucleic Acids Res. 40, 8276–8284.
Shen M.R., Jones I.M., Mohrenweiser H. 1998. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 58, 604‒608.
Mohrenweiser H.W., Jones I.M. 1998. Variation in DNA repair is a factor in cancer susceptibility: A paradigm for the promises and perils of individual and population risk estimation? Mutat. Res. 400, 5‒24.
Hung R.J., Hall J., Brennan P., Boffetta P. 2005. Genetic polymorphisms in the base excision repair pathway and cancer risk: A huge review. Am. J. Epidemiol. 162, 925‒942.
Nohmi T., Kim S.R., Yamada M. 2005. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat. Res. 591, 60‒73.
Illuzzi J.L., Harris N.A., Manvilla B.A., Kim D., Li M., Drohat A.C., Wilson D.M. 2013. Functional assessment of population and tumor-associated APE1 protein variants. PLoS One. 8, e65922.
Kim W.C., Ma C., Li W.M., Chohan M., Wilson D.M., Lee C.H. 2014. Altered endoribonuclease activity of apurinic/apyrimidinic endonuclease 1 variants identified in the human population. PLoS One. 9, 1‒9.
Kwiatkowski D., Czarny P., Galecki P., Bachurska A., Talarowska M., Orzechowska A., Bobinska K., Bielecka-Kowalska A., Pietras T., Szemraj J., Maes M., Sliwinski T. 2015. Variants of base excision repair genes MUTYH, PARP1 and XRCC1 in Alzheimer’s disease risk. Neuropsychobiology. 71, 176–186.
Czarny P., Kwiatkowski D., Toma M., Kubiak J., Sliwinska A., Talarowska M., Szemraj J., Maes M., Galecki P., Sliwinski T. 2017. Impact of single nucleotide polymorphisms of base excision repair genes on DNA damage and efficiency of DNA repair in recurrent depression disorder. Mol. Neurobiol. 54, 4150–4159.
Marsden C.G., Dragon J.A., Wallace S.S., Sweasy J.B. 2017. Base excision repair variants in cancer. Methods Enzymol. 591, 119–157.
Chan K.K.L., Zhang Q.M., Dianov G.L. 2006. Base excision repair fidelity in normal and cancer cells. Mutagenesis. 21, 173‒178.
Sweasy J.B., Lang T.M., DiMaio D. 2006. Is base excision repair a tumor suppressor mechanism? Cell Cycle. 5, 250–259.
Tudek B. 2007. Base excision repair modulation as a risk factor for human cancers. Mol. Aspects Med. 28, 258‒275.
D’Errico M., Parlanti E., Dogliotti E. 2008. Mechanism of oxidative DNA damage repair and relevance to human pathology. Mutat. Res. 659, 4‒14.
Nemec A.A., Wallace S.S., Sweasy J.B. 2010. Variant base excision repair proteins: contributors to genomic instability. Semin. Cancer Biol. 20, 320‒328.
Wilson D.M., Kim D., Berquist B.R., Sigurdson A.J. 2011. Variation in base excision repair capacity. Mutat. Res. 711, 100‒112.
Wallace S.S., Murphy D.L., Sweasy J.B. 2012. Base excision repair and cancer. Cancer Lett. 327, 73‒89.
Karahalil B., Bohr V.A., Wilson D.M. 2012. Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk. Hum. Exp. Toxicol. 31, 981‒1005.
Shapovalov M.V., Dunbrack R.L. 2011. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 19, 844–858.
Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. 1995. A second generation all atom force field for the simulation of proteins, nucleic acids and organic molecules. J. Am. Chem. Soc. 117, 5179–5197.
Wang J., Cieplak P., Kollman P.A. 2000. How well does a Restrained Electrostatic Potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21, 1049–1074.
Popov A.V., Vorob’ev Yu.N. 2010. GUI-BioPASED: A program for molecular dynamics simulations of biopolymers with a graphical user interface. Mol. Biol. (Moscow). 44 (4), 648–654.
Lazaridis T., Karplus M. 1999. Effective energy function for proteins in solution. Proteins. 35, 133–152.
Kuznetsova A.A., Kuznetsov N.A., Ishchenko A.A., Saparbaev M.K., Fedorova O.S. 2014. Pre-steady-state fluorescence analysis of damaged DNA transfer from human DNA glycosylases to AP endonuclease APE1. Biochim. Biophys. Acta. 1840, 3042–3051.
Yakovlev D.A., Kuznetsova A.A., Fedorova A.S., Kuznetsov N.A. 2017. Search for modified DNA sites with the human methyl-CpG-binding enzyme MBD4. Acta Naturae. 9, 95–105.
Petronzelli F., Riccio A., Markham G.D., Seeholzer S.H., Genuardi M., Karbowski M., Yeung A.T., Matsumoto Y., Bellacosa A. 2000. Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): Fundamental role of the catalytic domain. J. Cell Physiol. 185, 473–480.
Funding
This work was supported by grant no. 16-14-10038 from the Russian Science Foundation and partial support from budgetary funding (no. АААА-А17-117020210022-4) to ensure routine maintenance of the equipment.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare they have no conflict of interest. This study does not contain any research involving humans or animals as research objects.
Rights and permissions
About this article
Cite this article
Alekseeva, I.V., Bakman, A.S., Iakovlev, D.A. et al. Comparative Analysis of the Activity of the Polymorphic Variants of Human Uracil-DNA-Glycosylases SMUG1 and MBD4. Mol Biol 55, 241–251 (2021). https://doi.org/10.1134/S0026893321020035
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893321020035