Abstract—
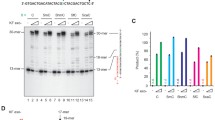
5-Methyl-2'-deoxycytidine (mC) and the product of its controlled oxidation, 5-hydroxymethyl-2'-cytidine (hmC), play a key role in the epigenetic regulation of gene expression, the cell differentiation, and the carcinogenesis. Due to spontaneious deamination, genomic CpG sites containing mC and hmC serve as mutagenesis hotspots. In addition, error-prone translesion and reparative DNA polymerases may serve as additional source of mutations in the lesion-containing regions with CpG sites. In the present work, we performed in vitro analysis of the accuracy of nucleotide incorporation opposite to mC and hmC by human DNA polymerases Polβ, Polλ, Polη, Polι, Polκ and primase polymerase PrimPol. The results of the study show a high accuracy of copying mC and hmC by the reparative DNA polymerases Polβ and Polλ, while Polη, Polι, Polκ, and PrimPol copied mC and hmC with less accuracy evident by incorporation of dAMP and dTMP. The same spectrum of error-prone dNMP incorporation was also noted at sites with unmodified cytosines.


Similar content being viewed by others
REFERENCES
Weber M., Schübeler D. 2007. Genomic patterns of DNA methylation: Targets and function of an epigenetic mark. Curr. Opin. Cell Biol. 19, 273–280.
Law J.A., Jacobsen S.E. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220.
Szulwach K.E., Jin P. 2014. Integrating DNA methylation dynamics into a framework for understanding epigenetic codes. Bioessays. 36, 107–117.
Jabbari K., Bernardi G. 2004. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene. 333, 143–149.
Stevens M., Cheng J.B., Li D., Xie M., Hong C., Maire C.L., Ligon K.L., Hirst M., Marra M.A., Costello J.F., Wang T. 2013. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 23, 1541–1553.
Branco M.R., Ficz G., Reik W. 2011. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13.
Shen L., Zhang Y. 2013. 5-Hydroxymethylcytosine: Generation, fate, and genomic distribution. Curr. Opin. Cell Biol. 25, 289–296.
Lu X., Zhao B.S., He C. 2015. TET family proteins: Oxidation activity, interacting molecules, and functions in diseases. Chem. Rev. 115, 2225–2239.
Wu X., Zhang Y. 2017. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534.
Waters T.R., Swann P.F. 2000. Thymine-DNA glycosylase and G to A transition mutations at CpG sites. Mutat. Res. 462, 137–147.
Pfeifer G.P. 2006. Mutagenesis at methylated CpG sequences. Curr. Top. Microbiol. Immunol. 301, 259–281.
Olinski R., Starczak M., Gackowski D. 2016. Enigmatic 5-hydroxymethyluracil: oxidatively modified base, epigenetic mark or both? Mutat. Res. 767, 59–66.
Kawai K., Wata Y., Hara M., Tojo S., Majima T. 2002. Regulation of one-electron oxidation rate of guanine by base pairing with cytosine derivatives. J. Am. Chem. Soc. 124, 3586–3590.
Denissenko M.F., Pao A., Tang M.-S., Pfeifer G.P. 1996. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 274, 430–432.
Hu W., Feng Z., Tang M.-S. 2003. Preferential carcinogen-DNA adduct formation at codons 12 and 14 in the human K-ras gene and their possible mechanisms. Biochemistry. 42, 10012–10023.
Vairapandi M., Duker H.J. 1994. Excision of ultraviolet-induced photoproducts of 5-methylcytosine from DNA. Mutat. Res. 315, 85–94.
You Y.-H., Li C., Pfeifer G.P. 1999. Involvement of 5‑methylcytosine in sunlight-induced mutagenesis. J. Mol. Biol. 293, 493–503.
Lee D.-H., Pfeifer G.P. 2003. Deamination of 5-methylcytosines within cyclobutane pyrimidine dimers is an important component of UVB mutagenesis. J. Biol. Chem. 278, 10314–10321.
Tomkova M., McClellan M., Kriaucionis S., Schuster-Böckler B. 2018. DNA replication and associated repair pathways are involved in the mutagenesis of methylated cytosine. DNA Repair (Amst.). 62, 1–7.
Tomkova M., Schuster-Böckler B. 2018. DNA modifications: Naturally more error prone? Trends Genet. 34, 627–638.
Yamtich J., Sweasy J.B. 2010. DNA polymerase family X: Function, structure, and cellular roles. Biochim. Biophys. Acta. 1804, 1136–1150.
Pata J.D. 2010. Structural diversity of the Y-family DNA polymerases. Biochim. Biophys. Acta. 1804, 1124–1135.
Yang W. 2014. An overview of Y-family DNA polymerases and a case study of human DNA polymerase η. Biochemistry. 53, 2793–2803.
Makarova A.V., Burgers P.M. 2015. Eukaryotic DNA polymerase ζ. DNA Repair (Amst.). 29, 47–55.
García-Gómez S., Reyes A., Martínez-Jiménez M.I., Chocrón E.S., Mourón S., Terrados G., Powell C., Salido E., Méndez J., Holt I.J., Blanco L. 2013. PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell. 52, 541–553.
Belousova E.A., Lavrik O.I. 2010. DNA polymerases beta and lambda, and their roles in the DNA replication and repair. Mol. Biol. (Moscow). 44, 839–855.
Lindahl T., Barnes D.E. 2000. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 65, 127–133.
Maga G., Villani G., Crespan E., Wimmer U., Ferrari E., Bertocci B., Hübscher U. 2007. 8-Oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 447, 606–608.
Makarova A.V., Grabow C., Gening L.V., Tarantul V.Z., Tahirov T.H., Bessho T., Pavlov Y.I. 2011. Inaccurate DNA synthesis in cell extracts of yeast producing active human DNA polymerase iota. PLoS One. 6, e16612.
Makarova A.V., Stodola J.L., Burgers P.M. 2012. A four-subunit DNA polymerase ζ complex containing Polδ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 40, 11618–11626.
Makarova A.V., Nick McElhinny S.A., Watts B.E., Kunkel T.A., Burgers P.M. 2014. Ribonucleotide incorporation by yeast DNA polymerase ζ. DNA Repair (Amst.). 18, 63–67.
Boldinova E.O., Stojkovič G., Khairullin R., Wanrooij S., Makarova A.V. 2017. Optimization of the expression, purification and polymerase activity reaction conditions of recombinant human PrimPol. PLoS One. 12, e0184489.
Petrova D.V., Naumenko M.B., Khantakova D.V., Grin I.R., Zharkov D.O. 2019. Relative efficiency of recognition of 5-methylcytosine and 5-hydroxymethylcytosine by methyl-dependent DNA endonuclease GlaI. Russ. J. Bioorg. Chem. 45, 625–629.
Johnson R.E., Washington M.T., Prakash S., Prakash L. 2000. Fidelity of human DNA polymerase η. J. Biol. Chem. 275, 7447–7450.
Zhang Y., Yuan F., Wu X., Wang Z. 2000. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase ι. Mol. Cell. Biol. 20, 7099–7108.
Zhang Y., Yuan F., Xin H., Wu X., Rajpal D.K., Yang D., Wang Z. 2000. Human DNA polymerase κ synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 28, 4147–4156.
Boldinova E.O., Wanrooij P.H., Shilkin E.S., Wanrooij S., Makarova A.V. 2017. DNA damage tolerance by eukaryotic DNA polymerase and primase PrimPol. Int. J. Mol. Sci. 18, 1584.
Poulos R.C., Olivier J., Wong J.W.H. 2017. The interaction between cytosine methylation and processes of DNA replication and repair shape the mutational landscape of cancer genomes. Nucleic Acids Res. 45, 7786–7795.
Rogozin I.B., Goncearenco A., Lada A.G., De S., Yurchenko V., Nudelman G., Panchenko A.R., Cooper D.N., Pavlov Y.I. 2018. DNA polymerase η mutational signatures are found in a variety of different types of cancer. Cell Cycle. 17, 348–355.
Tomkova M., Tomek J., Kriaucionis S., Schuster-Böckler B. 2018. Mutational signature distribution varies with DNA replication timing and strand asymmetry. Genome Biol. 19, 129.
Fang H., Barbour J.A., Poulos R.C., Katainen R., Aaltonen L.A., Wong J.W.H. 2020. Mutational processes of distinct POLE exonuclease domain mutants drive an enrichment of a specific TP53 mutation in colorectal cancer. PLoS Genet. 16, e1008572.
Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.-L., Boyault S., Burkhardt B., Butler A.P., Caldas C., Davies H.R., et al. 2013. Signatures of mutational processes in human cancer. Nature. 500, 415–421.
Alsøe L., Sarno A., Carracedo S., Domanska D., Dingler F., Lirussi L., SenGupta T., Tekin N.B., Jobert L., Alexandrov L.B., Galashevskaya A., Rada C., Sandve G.K., Rognes T., Krokan H.E., Nilsen H. 2017. Uracil accumulation and mutagenesis dominated by cytosine deamination in CpG dinucleotides in mice lacking UNG and SMUG1. Sci. Rep. 7, 7199.
ESCODD (European Standards Committee on Oxidative DNA Damage). 2002. Comparative analysis of baseline 8-oxo-7,8-dihydroguanine in mammalian cell DNA, by different methods in different laboratories: an approach to consensus. Carcinogenesis. 23, 2129–2133.
ESCODD (European Standards Committee on Oxidative DNA Damage), Gedik C.M., Collins A. 2005. Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 19, 82–84.
ACKNOWLEDGMENTS
We are grateful to L.V. Gening (Institute of Molecular Genetics) for the Pol κ preparation, O.I. Lavrik (Institute of Chemical Biology and Fundamental Medicine) for the Polλ expression vector, and K.A. Bondarenko for help in Polη purification.
Funding
This work was supported by the Russian Foundation for Basic Research (project nos. 17-00-00264-komfi (AVM) and 17-00-00261-komfi (DOZ)). PrimPol activity testing was supported by the Russian Science Foundation (project no. 18-14-00354 (AVM)).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This work does not contain any studies involving animals or human subjects performed by any of the authors.
Additional information
Translated by T. Tkacheva
Abbreviations: mC, 5-methyl-2'-deoxycytidine; hmC, 5-hydro-xymethyl-2'-deoxycytidine; oxoG, 8-oxo-7,8-dihydro-2'-deoxy-guanosine.
Rights and permissions
About this article
Cite this article
Shilkin, E.S., Petrova, D.V., Poltorachenko, V.A. et al. Template Properties of 5-Methyl-2'-Deoxycytidine and 5-Hydroxymethyl-2'-Deoxycytidine in Reactions with Human Translesion and Reparative DNA Polymerases. Mol Biol 55, 267–272 (2021). https://doi.org/10.1134/S0026893321020138
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893321020138