Microbial Communities of the Drinking Water With Gradient Radon Concentration Are Primarily Contributed by Radon and Heavy Metal Content
- 1Department of Life Science and Biotechnology, Jadavpur University, Kolkata, India
- 2Department of Physics, School of Studies in Environmental Radiation and Archaeological Sciences, Jadavpur University, Kolkata, India
Radon and heavy metal (HM) contamination in drinking water and their impact on health have been reported earlier. However, relatively little is known about the microbial community in drinking water with gradients of radon and the drivers of microbial community patterns in such water. With this view, we first examine microbial dynamics of drinking water in the permissible level of 93 ± 2 Bq/l as control, 510 ± 1.5 6 Bq/l and 576 ± 2 Bq/l as medium, and 728 ± 3 Bq/l as high radon-containing tube wells from Dumka and Godda districts, which comes under a major fault of the eastern fringes of India. Attempts have also been made to predict the impact of the radon contamination gradient and other water environmental parameters on community structure. The measured physicochemical character revealed strong clustering by the sampling site with respect to its radon and HM content. The radon-contaminated sites represent HM-rich nutrient-limited sites compared to the control. Radon (Rn), HM (Pb, Cu, and As), and total suspended solids (TSSs) were the most determinant variable among the parameters and influenced the microbial community composition of that region. The microbial diversity of those sites was lower, and this measured diversity decreased gradually on the sites with an increased gradient of radon contamination. The dominant microbial families in the contaminated sites were Moraxellaceae, Chitinophagaceae, unclassified Candidatus Azambacteria, unclassified Candidatus Moranbacteria, unclassified Candidatus Collierbacteria, and Gammaproteobacterial members, which are reported to abundantly inhabit radiation and chemolithotrophic environments and pose better radionuclide protective mechanisms, while the bacterial members dominant in the control site were Comamonadaceae, Rhodocyclaceae, Nitrospirales Incertae Sedis, cvE6, unclassified Woesearchaeota (DHVEG-6), and Holophagaceae, which are reported to be abundant in natural soil and drinking water, and labile in harsh environments. Relative sequence abundance of Comamonadaceae was decreasing on the sites with an increasing radon gradient, while the opposite trend was observed for Chitinophagaceae. The distribution of such microbial assemblages is linked to radon and heavy metal, highlighting that taxa with distinct environmental preferences underlie apparent clustering by sites; thus, we can utilize them for biostimulation-based in situ bioremediation purposes.
Introduction
Radon (222Rn) is a water-soluble, colorless, and odorless noble gas produced via alpha decay of radium (226Ra) in uranium (238U) decay series, which emits an alpha particle (Somlai et al., 2007). Long-term radon exposure can cause serious health issues. It is the second leading reason for lung cancer, followed by tobacco smoking (Thompson, 2011; Binesh et al., 2012). Radon continuously enters the water from radionuclide-containing rocks of the terrestrial crust and migrates with groundwater in large areas due to its water solubility (Naskar et al., 2018). According to the EU Commission and the WHO, the proposed permissible limit for drinking water is 100 Bq/l, and the lowest maximum contamination level (MCL) of drinking water is 11.1 Bq/l, as proposed by the U.S. Environmental Protection Agency (Naskar et al., 2018). It is frequently observed that radon-containing groundwater is also co-contaminated with other radionuclides and toxic heavy metals (HM) (Majumdar et al., 2000). The population living on the surface of those areas use such toxic water for their drinking and all other domestic purposes, which may be associated with carcinogenic risk and frequent death (Al-Zoughool and Krewski, 2009).
While most of the higher organisms are labile against this toxicity of radon and other radionuclides, the inhabitant microorganisms show resistance due to their protective mechanisms that are provided by extremozymes and extremolytes (Shukla et al., 2007). The radiation survival pure culture may not be effective in the relatively nutrient-limited different amounts of radiation-contaminated environments for bioremediation purposes due to community competition and other selective stress (Brown et al., 2015). However, indigenous microbial-based biostimulation or bioaugmentation is a more efficient approach than others that can be exploiting (Hassanshahian et al., 2014; Shukla et al., 2017). Therefore, characterization of these indigenous microbial communities from a gradient dose of radon-contaminated environment is extremely important to decipher their mode of interaction with radon, HM, and other environmental parameters to formulate effective bioremediation strategies. In this regard, the changes of the microbial community, if any, with respect to gradient concentration of the contaminants, also need to be addressed.
There are several reports on the measurement of radon concentration in groundwater, hot springs, soil, thermal plants, and indoor air. Attempts have also been made to correlate earthquakes with the radon level in groundwater and soil (Negarestani et al., 2002; Darby et al., 2005; Singh et al., 2008; Kansal et al., 2012). However, the microbiology of radon-contaminated drinking water has not been researched yet. Nevertheless, this is the first report on the microbial community composition of radon- and HM-contaminated drinking water. In this investigation, the samples were collected from already reported radon- and HM-contaminated drinking water tube wells from Dumka and Godda districts of Jharkhand, which is the part of Son–Narmada–Tapti, a major fault of the eastern fringes of India. It was reported that more than 42% of the drinking water tube wells of this region are highly contaminated with radon and HM (Naskar et al., 2018; Nayak et al., 2020). That contaminated water is being used for drinking and other domestic purposes by the native population. However, the aim of this investigation was as follows: 1) characterization of the sampling sites based on their major environmental parameters, 2) phylogenetics of the indigenous microbial community (based on 16S rRNA marker gene amplicon sequencing on the Illumina MiSeq platform) in different drinking water samples with gradient contamination of radon, and 3) evaluation of the role of measured environmental parameters for their influential role in community composition.
Materials and Methods
Site Description and Sample Collection
Groundwater was collected in July 2016 from four different deep tube wells with a depth of 24–76 m based on previously reported radon contamination (Naskar et al., 2018). Among the sampling sites, Station-4 (S4) was reported to have the highest radon concentration, Station-3 (S3) and Station-2 (S2) were reported to have medium radon contamination, and Station-1 (S1), which came under the permissible dose of radon in drinking water, was considered in the study for better comparison. After pumping the tube wells for 5 min, samples were collected in autoclaved bottles, and the bottles were overfilled and then capped under the water to avoid air bubbles. All the samples were collected in triplicate and brought to the laboratory within 4°C insulated chambers for further study. Therefore, a total of twelve samples, three from each of the four sites, were considered for this investigation, and the coordinates of the sampling sites were placed on the map (Figure 1).
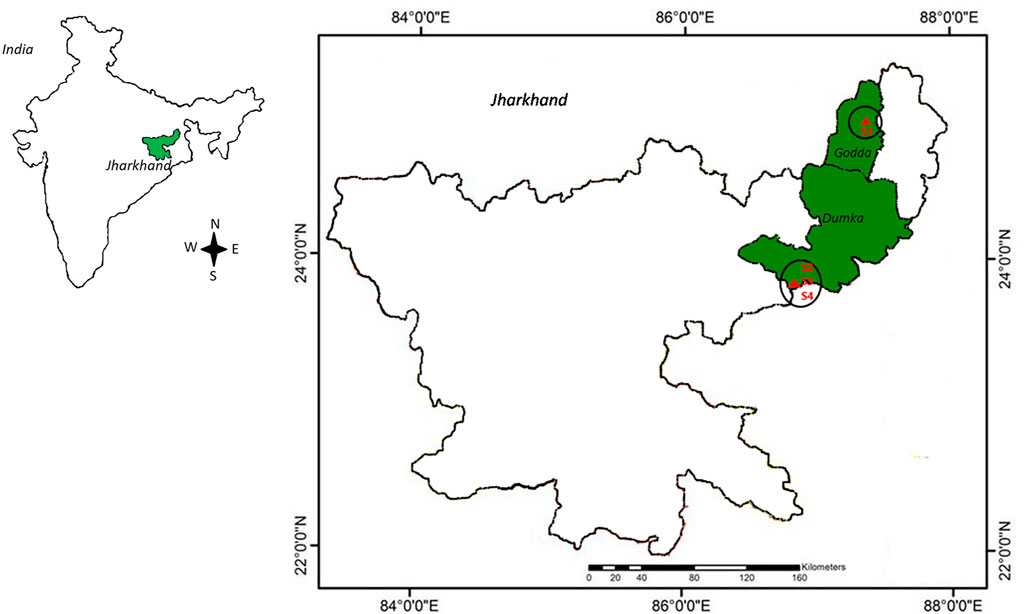
FIGURE 1. Map of the sampling sites. The permissible level of radon-contaminated drinking water (S1) was collected from Godda, whereas medium (S2 and S3) and highly contaminated water (S4) was taken from Dumka district of Jharkhand, India. The GPS data of sampling sites were compiled in QGIS software for mapping (https://www.qgis.org/en/site/).
Water Physiochemical Parameter
The water temperature and pH were measured in situ by using a mercury thermometer and a pH meter (Thermo Scientific™ Orion Star™ A211), respectively. Radon concentration was analyzed on the spot by using AlphaGuard portable radon monitor systems along with an AquaKit (Instruments G, Germany) (Naskar et al., 2018). Heavy metal (Zn, As, Cu, and Pb) concentration was measured using an atomic absorption spectrophotometer (AAS) (PerkinElmer, Analyst 300, United States), while Cr concentration was measured colorimetrically (Rice et al., 2017). Nutrient contents were also analyzed, which included total nitrogen (TN) (Macro-Kjeldahl method), total phosphate (TPH) (stannous chloride method), total potassium (TP) (flame photometric method), total suspended solids (TSSs) (total suspended solids were dried), and volatile suspended solids (VSSs) (fixed and volatile solids were ignited), guided by Rice et al. (2017) (Supplementary Table S2).
Meta-Genomic DNA Extraction and Illumina Amplicon Sequencing
One liter of water from each of the samples was filtered aseptically using a 0.22 µ filter paper. Those filter papers were further used for meta-genomic extraction using a PowerWater® DNA Isolation Kit (MO BIO), and DNA concentration was determined by using a Qubit 4 fluorometer (ThermoFisher SCIENTIFIC). The hypervariable V3–V4 regions of the bacterial 16S rRNA gene were amplified from the extracted metagenomic DNA samples using universal 16S rRNA primer (Dhal et al., 2020) to generate an amplicon library using the Nextera XT Index kit (Illuminainc) and the Nextera XT DNA Library Prep Kit (Part # 15044223 Rev. B). Next, the amplicon sequencing data were generated on the Illumina MiSeq platform in a 2 × 300 bp paired-end run. The primer-trimmed raw paired-end sequences were provided by Eurofins, India.
Sequence Processing
All the raw fastq datasets were processed using the following sequence processing protocol (Hassenrück et al., 2016). The paired-end amplicon sequences were trimmed based on a minimum quality filter score of 15 and a window size of 4 bases using trimmomatic v0.32 (Bolger et al., 2014). Then trimmed sequences were merged using PEAR v0.9.5 (Zhang et al., 2014), and OTU (operational taxonomic unit) clustering was performed using swarm v2.0 with the default value (Mahé et al., 2014). Sequence OTUs were taxonomically classified using SINA v1.2.11 and SILVA rRNA project reference database 128 with a minimum similarity alignment of 0.9 (Pruesse et al., 2012). The obtained OTUs were further curated to exclude the absolute singleton (occurring only once in the complete dataset), mitochondria/chloroplast DNA, and unclassified sequences on the phylum level using well-standardized R scripts (Hassenrück et al., 2016).
Statistical Analysis
All statistical analyses were conducted in the core distribution of R v3.4.2 (R Core Team, 2017) and R-Studio, v1.0.153 (RStudio Team, 2017). A principal component analysis (PCA) was performed to perceive patterns among the samples based on their environmental characteristics. Temperature and pH were excluded in this analysis as their values did not significantly differ between sample sites. Volatile suspended solids were also excluded because they were covered up within the total suspended solids. Variations in the water parameter between the contaminated and control sites were measured by two-way ANOVA, and correlation of radon with other physiochemical parameters was analyzed through the Spearman rank correlation test (Nayak et al., 2020).
The α-diversity indices were calculated based on repeated random subsampling of the amplicon datasets after randomly rarefying the data. To assess the richness and evenness, OTU number, abundance-based coverage estimator, inverse Simpson index, and Shannon diversity index were calculated. The significant difference in species richness between the sites was measured by the Kruskal–Wallis test. The relationship between α-diversity and water parameters (if any) was also assessed by the Spearman rank correlation test.
For the amplicon datasets, we excluded the rare biosphere by retaining OTUs that were present in more than two sequences in more than 10% of the samples. This reduction of the datasets did not change β-diversity patterns (Mantel test; r = 0.99, p < 0.001). Permutational multivariate analysis of variance (PERMANOVA) was calculated to check the differences in community-level between radon-contaminated and non-contaminated sites. The changes in community structure (β-diversity) between the studied sites were explored mainly by calculating the Bray–Curtis dissimilarity matrix using their respective relative OTU abundance datasets. This was used for nonmetric multidimensional scaling (NMDS) plot to visualize the patterns in the bacterial community composition for each habitat, and an envfit model was used for correlation pattern analysis within the water parameters and microbial community. The length of the arrow is proportional to the correlation between ordination and environmental variable.
The contribution of environmental parameters to explaining the variation in community structure redundancy analysis (RDA) was calculated using centered log-transformed relative OTU abundances. Before significance testing, parameters were excluded using forward model selection until the minimum Akaike information criterion (AIC), and the maximum adjusted R2 value was reached. Sequence counts were clr-transformed with aldex.clr function with a median of 128 Monte Carlo Dirichlet using the ALDEx2 package. The differentially abundant OTU among the radon-contaminated and non-contaminated sites was reflected in Dotplot prior to this test.
All statistical analyses and figure visualizations were performed in R v3.4.2 (R Core Team, 2017) with the packages vegan (Oksanen et al., 2019) and ALDEx2 (Fernandes et al., 2014).
Nucleotide Accession Number
Raw sequences used in this study were submitted to the NCBI with the biosample accession numbers SRX5391781, SRX5391782, SRX5391783, and SRX5391784. Each of the biosamples represents each site, and within every biosample, three raw sequence reads were submitted. The direct link to the deposited data is https://www.ncbi.nlm.nih.gov/sra/PRJNA522844.
Results
Water Quality Parameters
In this investigation, sampling sites were selected based on the radon concentration gradient of the drinking water (Nayak et al., 2020). The radon concentration of site S1 was 93 ± 2 Bq/l, which lies within the permissible dose of radon (100 Bq/l) recommended by the WHO (2011), and it served as a control site. The samples of sites S2 and S3 contained medium concentrations (510 ± 1.5 and 576 ± 2 Bq/l, respectively), and later, site S4 contained the highest radon concentration (728 ± 3 Bq/l). Noteworthily, radon- and HM-contaminated sites S2, S3, and S4 varied significantly with respect to the control (ANOVA, p < 0.01). Based on our data, the nutrient contents of our studied samples were more or less similar to the groundwater nutrient content of New Jersey, where they mention such nutrient concentration as low, which may be designated as nutrient-limited environments (Nayak et al., 2020). The Spearman rank correlation test indicated that within the measured water parameters, Pb, Cr, Cu, As, and TSSs had a significant level of positive correlation with radon, while TP had negative correlation with radon (Supplementary Table S3A). Therefore, it was observed that along the radon gradient selected, HM (Pb, Cr, Cu, and As) and TSS concentration increased, while TP content decreased significantly. The PCA analysis indicated that the first two principal components accounted for 91.5% of the variation among the sites based on their measured water quality parameters (Figure 2). The PC1 which alone explained 73.2% of the variation was majorly contributed by the radon, HM (As, Cu, Cr, and Pb), TN, TP, and TSS content, while the PC2 responsible for the remaining 18.3% of the total variation was mainly derived from the Zn and TPH content of the sites. As indicated in Figure 2, the sampling sites were segregated into three (S1, S2, and S3 as a single cluster as they were placed nearby and S4) distinct clusters according to their radon concentration gradients. The contaminated and non-contaminated sites were separated along with the opposite orientation of the PC1 axis. The ordinations of contaminated sites were driven by radon and major heavy metals (As, Cu, Cr, and Pb), whereas the control site was driven by TN, TP, and TSSs (Figure 2; Supplementary Figure S1).
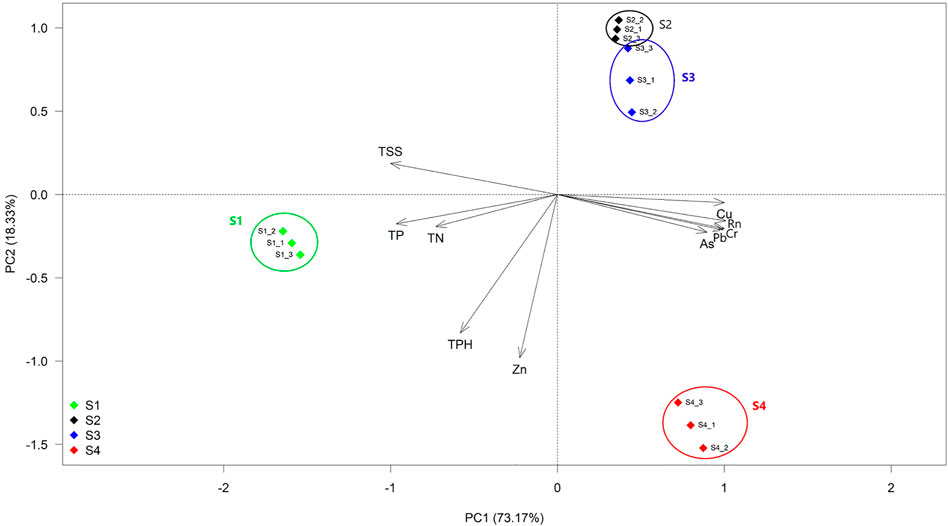
FIGURE 2. Principal component analysis (PCA) of water physiochemical parameters. The sample sites named S1 (blue), S2 (black), and S3 (red) are radon-contaminated, while S4 (green) is normal groundwater. PC1 and PC2 explained 73.17 and 18.33% of the total variation, respectively. Here, Rn: radon, and five heavy metals are As: arsenic, Cu: copper, Pb: lead, Cr: chromium, and Zn: zinc; TN: total nitrogen, TP: total potassium, TPH: total phosphate, and TSS: total suspended solid.
Diversity Analysis and Taxonomy Composition
A total of 85,32,914 clipped reads were generated after removing the primer sequences of the microbial hypervariable V3–V4 region of the 16S rRNA gene amplicon sequences. After removing absolute singletons and OTUs that classified to mitochondria/chloroplast DNA and unclassified sequences on the phylum level, a total of 54,813 swarm OTUs were obtained (Supplementary Table S1). Absolute singletons were removed in this study to reduce the rare biosphere, potential chimera effects, and PCR artifact (Hassenrück et al., 2016).
The α-diversity of each site was calculated based on the rarefied average OTUs per site, which ranged from 10,303 ± 356 to 8556 ± 301. Interestingly, species richness and evenness of the sites represented by abundance-based coverage estimator (invS), and Shannon diversity indices were decreasing with increasing gradient of radon contamination (Figure 3). This significant level of changes in species richness between the sites was confirmed by the Kruskal–Wallis test (χ2 = 9.4, df = 3, p-value < 0.05). The Spearman rank correlation test indicated that within the measured water parameters, radon, TSSs, and four other heavy metals (As, Cr, Pb, and Cu) significantly influenced the α-diversity (Supplementary Table S3B).
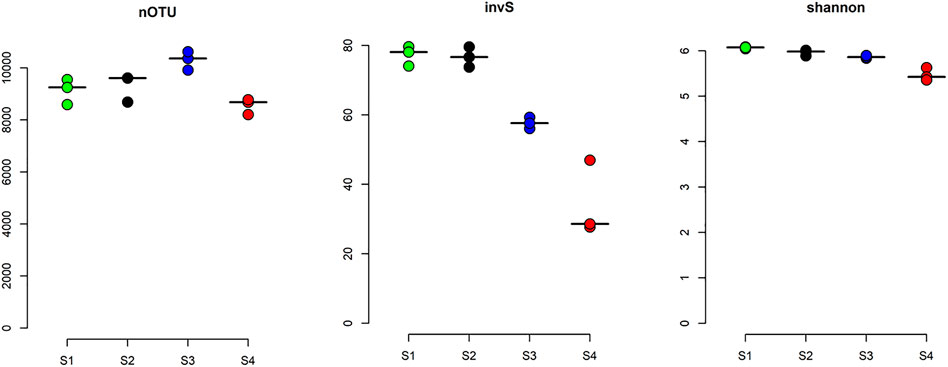
FIGURE 3. α-diversity of water samples. The α-diversity of the studied sites was measured based on their richness number of OTU (nOTU), inverse Simpson index (invS), and evenness (Shannon index). The lines here are corresponding median values.
The ten (R Core Team, 2017) most dominant sequences obtained in this study were affiliated to Proteobacteria (32–41% within all samples), Percubacteria (19–30%), Bacteriodetes (5–33%), and Nitrospirae (3–7%), followed by Chlamydiae, Microgenomates, Acitobacteria, Chloroflexi, Omnitrophica, Woesearchaeota (DHVEG–8), Euryarchaeota, Planctomycetes, Omnitrophica, Thaumarchaeota, and Ignavibacteria (Figure 5B). The relative sequence abundance of the bacterial phylum Nitrospirae showed decreasing trends (Spearman rank correlation; R = - 0.99, p-value < 0.01) in the sites with an increase in the concentration of radon, and a reversed result was observed for the phylum Bacteroidetes (Spearman rank correlation; R = - 0.71, p-value < 0.02). Among the dominant phyla, Proteobacteria, Parcubacteria, and Nitrospirae showed significant differences between the contaminated and non-contaminated sites (t-test, p < 0.01). In the lower taxonomy level, the relative sequence abundance affiliated to the family Comamonadaceae showed decreasing trends (Spearman rank correlation; R = - 0.80, p-value < 0.01) in the sites with a radon-contaminated gradient (S1 to S4), while a reversed order was observed for Chitinophagaceae (Spearman rank correlation; R = 0.98, p-value < 0.01; Figure 4). At the family level, a significant difference between the contaminated and control sites followed a similar result as described in their respective phylum levels.
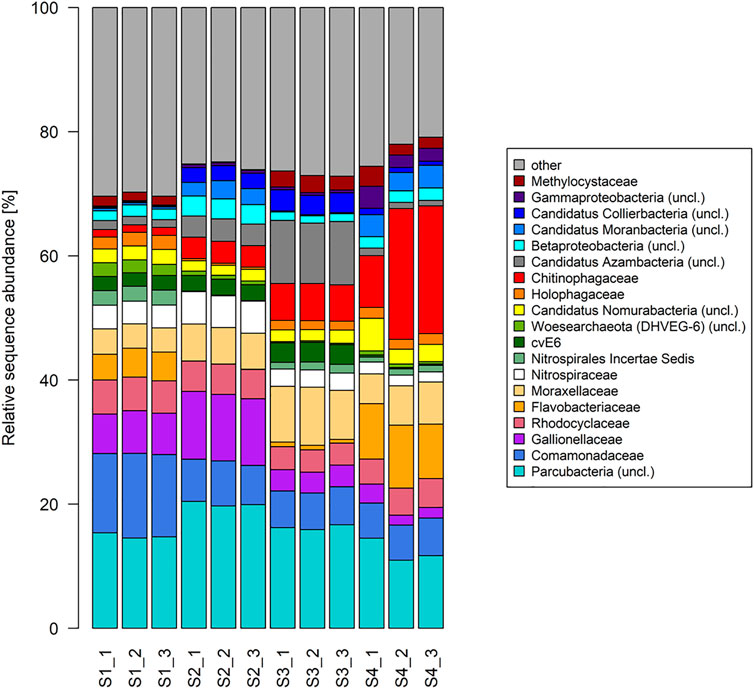
FIGURE 4. Taxonomic composition of radon-contaminated drinking water samples. Relative sequence abundance of 10 most dominant microbes in the family level of all samples.
At the OTU level, a significant difference in the bacterial community was observed between radon-contaminated and non-contaminated sites as indicated in PERMANOVA (R = 0.91, p = 0.001). This was confirmed by cluster analysis based on Bray–Curtis dissimilarities, which separate the contaminated (S4) and non-contaminated (S1) sites with an average of 0.85 dissimilarities (Figure 5A). To identify the OTUs responsible for this difference in community composition between the sampling sites, ALDEx2 was performed. A total of 52 OTUs representing Betaproteobacteria (9.7% of the total community), Parcubacteria (9%), Gammaproteobacteria (5.6%), Sphingobacteriia (5%), Flavobacteriia (3%), Alphaproteobacteria (2.7%), Candidatus Azambacteria (2.7%), Candidatus Nomurabacteria (1.5%), Candidatus Moranbacteria (1.5%), Candidatus Collierbacteria (1.5%), Nitrospira (1.3%), Chalamydiae (1%), Holophagae (0.6), Elusimicrobia (0.4%), and Omnitrophica Incertae Sedis (0.2%) were deferred as differential abundant between the radon-contaminated and non-contaminated sites (Figure 5C; Supplementary Figure S2). Within the Betaproteobacteria OTUs affiliated with Candidatus Nitrotoga (otu24), unclassified Comamonadaceae (otu25) was significantly enriched in contaminated sites, whereas Aquabacterium (otu6), Pseudorhodoferax (otu13), and unclassified Rhodocyclaceae (otu15) dominated in the control site. The unclassified genera Parcubacteria (otu4) was significantly dominant in contaminated sites. It was also observed that within the Gamaproteobacteria, Nitrospira, and Omnitrophica Incertae Sedis genera, unclassified Moraxellaceae (otu81), Candidatus Methylomirabillis (otu44), and Candidatus Omnitrophus (otu56) were significantly dominant in the non-contaminated site, respectively. Within the Alphaproteobacteria, the genera Rhizobium (otu36) showed higher abundance in contaminated sites, which varied significantly from the noncontaminated site. Similarly, among the class Chlamydiae, the Elusimicrobia, and Flavobacteriia genera, unclassified cvE6 (otu67), unclassified Lineage IV (otu42), and Chryseobacterium (otu18) significantly showed a higher presence in the non-contaminated site, respectively.
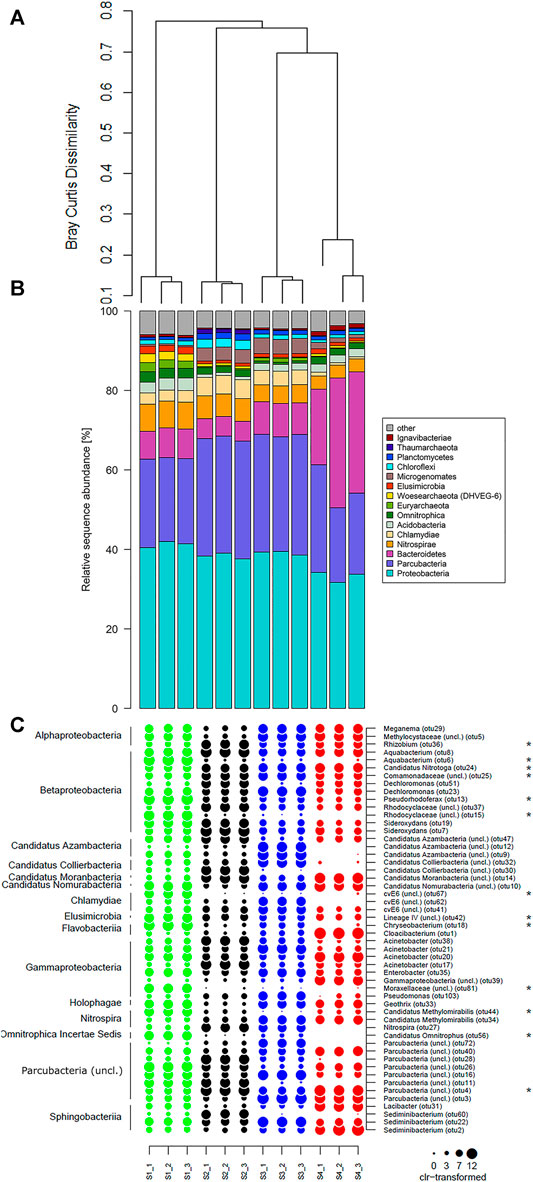
FIGURE 5. Dominant bacterial community of the investigated sites. Plots’ order from top to bottom: (A) hierarchical cluster dendrogram measured based on Bray–Curtis dissimilarity. (B) Taxonomy composition of 10 most dominant phyla with higher relative sequence proportions in bar plot. (C) Differentially abundant OTUs within four sites represented in Dotplot. Dotplot represents class-level taxonomy on the left side and genus-level taxonomy on the right side. The size of each dot (0, 5, 10, and 15) represents centered log ratio (clr)-transformed sequence counts.
According to dissimilarities, the sampling sites were projected in an ordination space and their associated environmental parameters on NMDS plots with a stress value < 0.01 (Figure 6). Based on this plot, contaminated communities were dissimilar to the non-contaminated site and were mainly associated with radon and HM, while the non-contaminated site was associated with nutrients of the water. The envfit result showed that Rn, TSSs, HM (Pb, Cu, and As), and TP coincided most strongly with the microbial community composition. Redundancy analysis, which was performed to assess the significant contribution of the tested parameters in describing the variations in bacterial communities, revealed that Rn, TSSs, HM (Pb, Cu, As, Cr, and Zn), TP, TN, and TPH explained 84% of the variation in bacterial communities (Table 1). Among the parameters, radon itself contributed 24.7%, followed by TSSs (24.5%), Pb (24.2%), Cu (24.2%), As (23.6%), and TP (22.7%), further confirming the NMDS plot ordination. Together, NMDS and RDA supported each other’s results and suggested that Rn, TSSs, Pb, Cu, and As were the most determinant variables among the parameters and showed dominancy in the explanation of the microbial community composition.
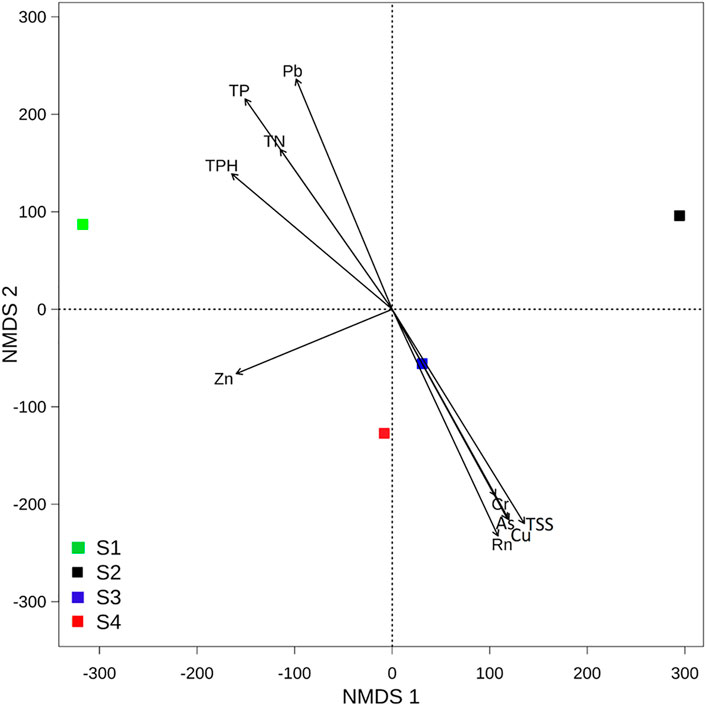
FIGURE 6. Nonmetric multidimensional scaling (NMDS) plot of the bacterial communities of each site (stress = 0.01). Arrows of the NMDS plot indicate envfit correlations of bacterial community composition with water quality parameters.
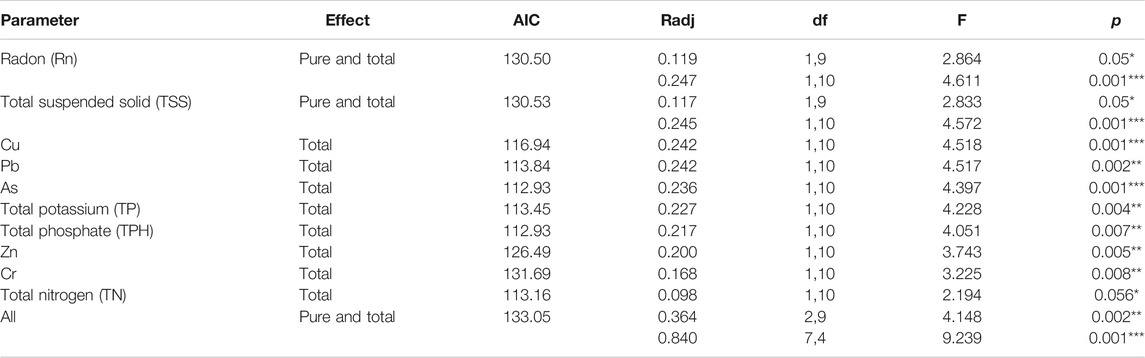
TABLE 1. Contribution of water parameters to variation in bacterial communities of studied groundwater. Total and pure effects (i.e., individual controlling factors of the analysis) of explanatory factors were calculated by using redundancy 610 analysis (RDA) models. The community variation is expressed by R2 as percentage values. For the significance test, the respective F-ratios were calculated by performing 1000 Monte Carlo permutation tests indicated by * = significant (p ≤ 0.05), ** = very significant (p ≤ 0.01), *** = highly significant (p ≤ 0.001), and not significant (p > 0.05). df: degrees of freedom (numerator, denominator).
Discussion
Radon contamination in the subsurface drinking water and its effects on human health were well investigated around the world, including India (Thabayneh, 2015; Naskar et al., 2018). But no report so far is available on the microbiology of such toxic water. In this context, this is the first attempt on the characterization of the indigenous microbial community in drinking water contaminated with radon and their community difference with increased radon levels with respect to control water from the same region, if any. In this respect, the aim of our study was to reveal the microbial community of radon-contaminated drinking water while trying to answer the following hypotheses: 1) radon-contaminated drinking water is a toxic, HM-rich, and nutrient-limited environment compared to normal water; 2) microbial richness and evenness change in the water with gradient radon concentration; 3) the composition of the microbial community differs with the radon contamination level; and 4) important environmental parameters including radon may play an influential role in community changes.
Defining the Sampling Sites Based on Measured Water Parameters
Sampling sites are selected from Dumka and Godda districts of Jharkhand, which is a part of a major fault of Son–Narmada–Tapti, India. Average radon contamination in the groundwater of this region is 106.8 Bq/l, higher than that in Extremadura, Spain (98 Bq/l), but below that in other regions of India like Garhwal (510 Bq/l) and Jaduguda (123.5 Bq/l) (Naskar et al., 2018). It is significantly above the safe limit of drinking water radon (WHO, 2011). The contaminated samples considered in this investigation showed a significantly elevated amount of radon and HM but a lower amount of nutrient than the control sample (ANOVA, p < 0.01). Noteworthily, the radon and HM concentrations of the water samples collected from contaminated sites are much higher, while the nutrient conditions are lower than their respective permissible limits in drinking water as proposed by the Bureau of Indian Standard (BIS) and the WHO (Chaurasia et al., 2018). Heavy metals in our water samples have a higher value than those in previously reported hydrocarbon- and radionuclide-contaminated groundwater (Reimann et al., 1996; Abbai and Pillay, 2013) and a lower level of nitrogen, phosphate, potassium, and dissolved solids than other radon-contaminated groundwater (Idriss et al., 2011). Within the observed water parameters, radon, HM (As, Cu, Cr, and Pb), and nutrients (TN, TP, and TSSs) show dominant explanatory power with regard to the environmental condition of that region and support the differentiation of the sampling sites as contaminated and non-contaminated, respectively. The major contaminants in the groundwater are the main driving forces of groundwater quality (Yan et al., 2003), which supports our observation as well. Therefore, our study sites can be defined as nutrient-limited, and radon- and HM-contaminated environments.
Microbial Richness and Evenness With Respect to Gradient Radon Contamination
This investigation indicates a gradient decrease of microbial richness and evenness in the samples with an increasing gradient of radon contamination (Kruskal–Wallis test; p-value < 0.05). Based on α-diversity indices, we observed a more diverse microbial community in the control drinking water than in the water from radon-contaminated sites. Our finding is in line with the previously reported groundwater microbial community where they observed a lower microbial diversity in terms of reduced bacterial richness and evenness in contaminated groundwater than normal (Hemme et al., 2015). Probably, environmental stress including radon, HM, and limiting nutrients are conditions that influence the diversity of the microbial community and flourish some specific types of communities in contaminated water.
Microbial Community Comparison Between Contaminated and Non-Contaminated Sites
At the family level, significantly dominant sequences obtained from contaminated sites are Moraxellaceae (under Proteobacteria), Chitinophagaceae (Bacteroidetes), unclassified Candidatus Azambacteria (Parcubacteria), unclassified Candidatus Moranbacteria (Parcubacteria), unclassified Candidatus Collierbacteria (Microgenomates), and unclassified Gammaproteobacteria (Proteobacteria), while Comamonadaceae (Proteobacteria), Rhodocyclaceae (Proteobacteria), Nitrospirales Incertae Sedis (Nitrospirae), cvE6 (Chlamydiae), unclassified Woesearchaeota (DHVEG-6) (Woesearchaeota), and Holophagaceae (Acidobacteria) are abundant in control sites (t-test; p < 0.05). The bacterial assemblages mentioned above were found to be dominated in contaminated radon water and were reported to be abundantly inhabitant in radiation and chemolithotrophic environments (Poirel et al., 2008; Brown et al., 2015; Rao et al., 2016; Danczak et al., 2017), thus supporting our finding. On the other side, dominant families present in the control water were generally reported to be present in natural soil and drinking water (Li et al., 2016; Deja-Sikora et al., 2019). This difference in communities between the contaminated and control sites is supported by PERMANOVA (p = 0.001) and Bray–Curtis dissimilarity cluster analysis that separates the control site from the cluster of contaminated sites. This investigation reports that within the dominated bacterial phylum, Nitrospirae showed decreasing trends in the water with an increased concentration of radon, and a reversed result was observed for the phylum Bacteroidetes. Nitrospira is a chemolithotrophic nitrite-oxidizing bacterium that shows the highest phylogenetic diversity and widest environmental distribution but is majorly found in fresh water and salt water (Daims et al., 2016) probably because of its reduced survival and adaptation strategies with a higher radionuclide-contaminated water. Meanwhile, Bacteroidetes was previously reported to be dominant in uranium-contaminated soils and γ-irradiated sediments (Brown et al., 2015; Yan et al., 2016), so it must have better survival strategies in such harsh environments. It is also reported that Bacteroidetes utilizes the catalase gene in order to show a resistance phenotype against oxidative stress (Rocha et al., 1996). At the lower taxonomy level, the relative sequence abundance affiliated to the family Chitinophagaceae showed increasing trends in the sites with a radon-contaminated gradient. Their presence in radiation-contaminated sediments is well documented as they are reported to have catalase and oxidase genes for radiation-related oxidative stress reduction potential (Brown et al., 2015; Dahal et al., 2017).
In Dotplot analysis, out of a total of 52 differentially abundant OTUs, the OTUs that were significantly enriched in contaminated sites are affiliated to the genera Candidatus Nitrotoga, unclassified Comamonadaceae, unclassified Parcubacteria, and the genera Rhizobium. Nitrite-oxidizing Candidatus Nitrotoga are widespread in a natural environment (Kitzinger et al., 2018), while unclassified Comamonadaceae is a dominant member of ancient subsurface and groundwater microbial communities in sulfidic waters reported as potential nitrate reducers and sulfur oxidizers (Deja-Sikora et al., 2019). The bacterial member of Parcubacteria has been reported as a dominant uncultivable microbe in a radiation-contaminated groundwater environment with limited metabolic genes, supporting their presence in contaminated sites which were considered in this investigation as a radiation-rich nutrient-deprived environment (Wrighton et al., 2012; Brown et al., 2015). They are also known to have unique metabolic genome sequences for energy and nutrient conservation and sustain in nutrient-limited environments (Wrighton et al., 2012). The abundance of Rhizobium is also observed in the microbial mat of mine water contaminated with heavy metals and uranium (Drewniak et al., 2016). On the other hand, the bacterial members of the genera Aquabacterium, Pseudorhodoferax, unclassified Rhodocyclaceae, unclassified Moraxellaceae, Candidatus Methylomirabillis, Candidatus Omnitrophus, Lineage IV (Elusimicrobia), cvE6 (Chlamydiae), and Chryseobacterium are significantly enriched in the control site. Interestingly, most of the genera mentioned above are dominantly found in drinking water biofilm (Aquabacterium), normal groundwater (Pseudorhodoferax), hard-rock aquifers (unclassified Rhodocyclaceae), freshwater ponds (unclassified Moraxellaceae and Candidatus Methylomirabillis), and aquatic environments (cvE6, Lineage IV, and Chryseobacterium) (Kalmbach et al., 1999; Corsaro et al., 2002; Song et al., 2008; Herlemann et al., 2009; Wu et al., 2012; Chen et al., 2013; Ben Maamar et al., 2015; Kolinko et al., 2016).
The presence of microbial members like unclassified Parcubacteria, Pseudorhodoferax, Sideroxydans, Nitrospira, unclassified Gammaproteobacteria, unclassified Betaproteobacteria, unclassified Methylocystaceae, unclassified Comamonadaceae, and Candidatus Methylomirabilis is well reported in radionuclide-, HM-, and oil-contaminated environments (Gihring et al., 2011; Maleke et al., 2015; Shukla et al., 2017; Kitzinger et al., 2018). Their mechanisms of interactions with such metal and radionuclide environments are also well investigated (Gihring et al., 2011; Maleke et al., 2015; Shukla et al., 2017; Kitzinger et al., 2018). However, a few of the other genera, such as Aquabacterium, Chryseobacterium, Acinetobacter, unclassified Woesearchaeota (DHVEG-6), unclassified cvE6, Sediminibacterium, Dechloromonas, Cloacibacterium, and Lacibacter, are neither reported to be present in radionuclide- and HM-contaminated environments nor are their metabolism and mechanism of interaction investigated; therefore, they may be the unique microbial assemblages for the studied areas.
Groundwater usually contains low concentrations of organic carbon; microbial life there depends on oxidation and reduction of inorganic compounds for energy (Ben Maamar et al., 2015). Radiation is potentially lethal to organisms because cytoplasmic water radiolysis generates quantities of reactive oxygen species (e.g., HO−, H2O2, and O2−), which may react indiscriminately with essential biomolecules, such as nucleic acids, proteins, and lipids, causing damage (Daly et al., 2007). Radiation resistance bacteria express DNA repair proteins, reactive oxygen species scavenger molecules, and hydrolytic enzymes like oxidase, catalase, and hydrogen peroxide. Therefore, radiation and HM help in the enrichment of radiation-resistant, chemolithotrophic, and reactive oxygen hydrolytic enzyme-producing microbes so that they are sustained in contaminated sites over the inhabitant microbes from normal water. A few of the selected members of the microbial communities in the investigated sites from the family level, that is, Comamonadaceae, Rhodocyclaceae, and Pseudomonadaceae, unclassified Parcubacteria, Pseudorhodoferax, Sideroxydans, Nitrospira, unclassified Gammaproteobacteria, unclassified Betaproteobacteria, unclassified Methylocystaceae, unclassified Comamonadaceae, and Candidatus Methylomirabilis, are well reported to be abundant in radionuclide- and HM-contaminated environments, and they also pose a strong radionuclide- and HM-removing ability (Brown et al., 2015; Drewniak et al., 2016; Yan et al., 2016; Dahal et al., 2017; Kitzinger et al., 2018; Deja-Sikora et al., 2019); therefore, they can serve as a potential target for in situ bioremediation through bioaugmentation or biostimulation.
Influence of Environmental Parameters on the Microbial Community
It is not easy to assess the environmental impacts on the microbial community because some specific environmental parameters cannot explain the total community of a site, as it is the combined effect of all parameters (Li et al., 2016). Geochemical conditions, in particular the availability of electron donors and acceptors, are the major driver of microbial community composition and diversity in heavy metal–contaminated groundwater (Ben Maamar et al., 2015). Except TSSs, other toxic HMs and radon have a negative correlation with bacterial diversity. This effect reflects on the NMDS, envfit, and RDA results which show that Rn, TSSs, Pb, Cu, As, and TP coincided most strongly with the microbial community composition of the contaminated sites. Our observation is well supported by the previous report that the environmental nutrients such as total carbon, nitrogen, phosphate, pH, dissolved oxygen, and total suspended solids had strong impacts on the microbial community, and heavy metals and radiation have negative effects on environments and select some specific types of microbes (Arroyo et al., 2015; Brown et al., 2015; Jiang et al., 2016; Li et al., 2016; Hou et al., 2017). Our investigation is in line with the previous report that indicates shifting of the microbial community where Bacteroidetes is dominant in the sediments with increasing exposure to radiation (Brown et al., 2015). Together, NMDS and RDA supported each other’s results and suggested that Rn, TSSs, Pb, Cu, and As were the most determinant variables among the parameters and explained the microbial community composition.
Conclusion
In situ bioremediation through biostimulation of the indigenous microbial community is a better approach than culture-based bioremediation (i.e., bioaugmentation), where community competition and other environmental stress have a negative impact on the whole process. In such a prospect, total community characterization and determination of environmental condition is one of the most important steps to start with. From the present investigation with high-throughput next-generation sequencing of 16S rRNA gene–based microbial community study and environmental condition of the radon- and HM-contaminated drinking water, the following conclusions can be drawn.
(1) Radon-contaminated drinking water is also co-contaminated with HM and creates high-stress conditions.
(2) This environmental stress reduces the microbial diversity compared to that in the control and helps in the enrichment of specific microbial communities according to the contamination level.
(3) Among the environmental parameters, Rn, Pb, Cu, As, and TSSs are major driving forces and community explanatory parameters.
(4) The bacterial members of Moraxellaceae, Chitinophagaceae, unclassified Candidatus, unclassified Gammaproteobacteria, Comamonadaceae, Rhodocyclaceae, genus-level unclassified Parcubacteria, Pseudorhodoferax, Sideroxydans, Nitrospira, unclassified Gammaproteobacteria, unclassified Betaproteobacteria, unclassified Methylocystaceae, unclassified Comamonadaceae, and Candidatus Methylomirabilis are known for their environmental stress resistance potential, and can be utilized for biostimulation-based in situ bioremediation purposes.
(5) The genera Aquabacterium, Chryseobacterium, Acinetobacter, unclassified Woesearchaeota (DHVEG-6), unclassified cvE6, Sediminibacterium, Dechloromonas, Cloacibacterium, and Lacibacter are unique microbial members in the investigated sites from radon- and HM-contaminated environments.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
PKD, TN, PK, and AD designed experiments. TN performed the experiments. TN, DD, and PKD analyzed data and assisted in data interpretation. PKD and TN wrote the manuscript with support and input from all coauthors. All authors critically revised the article and gave their approval of the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to the Department of Life Science and Biotechnology, Jadavpur University, for technical support. The authors also thank Chiranjib Barman for helping during the sampling. Student fellowship was provided by the Department of Biotechnology, Ministry of Science and Technology (Grant No. DBT/2016/JU/732). The financial assistance was taken from Jadavpur University central grand R-II/479/19 (RUSA 2.0). All the reviewers are thanked for their efforts to improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.576400/full#supplementary-material
References
Abbai, N. S., and Pillay, B. (2013). Analysis of Hydrocarbon-Contaminated Groundwater Metagenomes as Revealed by High-Throughput Sequencing. Mol. Biotechnol. 54, 900–912. doi:10.1007/s12033-012-9639-z
Al-Zoughool, M., and Krewski, D. (2009). Health Effects of Radon: A Review of the Literature. Int. J. Radiat. Biol. 85, 57–69. doi:10.1080/09553000802635054
Arroyo, P., Sáenz de Miera, L. E., and Ansola, G. (2015). Influence of Environmental Variables on the Structure and Composition of Soil Bacterial Communities in Natural and Constructed Wetlands. Sci. Total Environ. 506-507, 380–390. doi:10.1016/j.scitotenv.2014.11.039
Ben Maamar, S., Aquilina, L., Quaiser, A., Pauwels, H., Michon-Coudouel, S., Vergnaud-Ayraud, V., et al. (2015). Groundwater Isolation Governs Chemistry and Microbial Community Structure along Hydrologic Flowpaths. Front. Microbiol. 6, 1457. doi:10.3389/fmicb.2015.01457
Binesh, A., Mowlavi, A. A., Mohammadi, S., and Mowlavi, A. A. (2012). Estimation of the Effective Dose from Radon Ingestion and Inhalation in Drinking Water Sources of Mashhad, Iran. Int. J. Radiat. Res. 2.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Brown, A. R., Boothman, C., Pimblott, S. M., and Lloyd, J. R. (2015). The Impact of Gamma Radiation on Sediment Microbial Processes. Appl. Environ. Microbiol. 81, 4014–4025. doi:10.1128/AEM.00590-15
Chaurasia, A. K., Pandey, H. K., Tiwari, S. K., Prakash, R., Pandey, P., and Ram, A. (2018). Groundwater Quality Assessment Using Water Quality Index (WQI) in Parts of Varanasi District, Uttar Pradesh, India. J. Geol. Soc. India 92, 76–82. doi:10.1007/s12594-018-0955-1
Chen, W.-M., Lin, Y.-S., Young, C.-C., and Sheu, S.-Y. (2013). Pseudorhodoferax Aquiterrae Sp. nov., Isolated from Groundwater. Int. J. Syst. Evol. Microbiol. 63, 169–174. doi:10.1099/ijs.0.039842-0
Corsaro, D., Venditti, D., and Valassina, M. (2002). New Chlamydial Lineages from Freshwater Samples. Microbiology 148, 343–344. doi:10.1099/00221287-148-2-343
Dahal, R. H., Chaudhary, D. K., and Kim, J. (2017). Rurimicrobium Arvi Gen. Nov., Sp. nov., a Member of the Family Chitinophagaceae Isolated from Farmland Soil. Int. J. Syst. Evol. Microbiol. 67, 5235–5243. doi:10.1099/ijsem.0.002452
Daims, H., Lücker, S., and Wagner, M. (2016). A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria. Trends Microbiol. 24, 699–712. doi:10.1016/j.tim.2016.05.004
Daly, M. J., Gaidamakova, E. K., Matrosova, V. Y., Vasilenko, A., Zhai, M., Leapman, R. D., et al. (2007). Protein Oxidation Implicated as the Primary Determinant of Bacterial Radioresistance. Plos Biol. 5, e92. doi:10.1371/journal.pbio.0050092
Danczak, R. E., Johnston, M. D., Kenah, C., Slattery, M., Wrighton, K. C., and Wilkins, M. J. (2017). Members of the Candidate Phyla Radiation Are Functionally Differentiated by Carbon- and Nitrogen-Cycling Capabilities. Microbiome 5, 112. doi:10.1186/s40168-017-0331-1
Darby, S., Hill, D., Auvinen, A., Barros-Dios, J. M., Baysson, H., Bochicchio, F., et al. (2005). Radon in Homes and Risk of Lung Cancer: Collaborative Analysis of Individual Data from 13 European Case-Control Studies. BMJ 330, 223–226. doi:10.1136/bmj.38308.477650.63
Deja-Sikora, E., Gołębiewski, M., Kalwasińska, A., Krawiec, A., Kosobucki, P., and Walczak, M. (2019). Comamonadaceae OTU as a Remnant of an Ancient Microbial Community in Sulfidic Waters. Microb. Ecol. 78, 85–101. doi:10.1007/s00248-018-1270-5
Dhal, P. K., Kopprio, G. A., and Gärdes, A. (2020). Insights on Aquatic Microbiome of the Indian Sundarbans Mangrove Areas. PLoS One 15, e0221543. doi:10.1371/journal.pone.0221543
Drewniak, L., Krawczyk, P. S., Mielnicki, S., Adamska, D., Sobczak, A., Lipinski, L., et al. (2016). Physiological and Metagenomic Analyses of Microbial Mats Involved in Self-Purification of Mine Waters Contaminated with Heavy Metals. Front. Microbiol. 7, 1252. doi:10.3389/fmicb.2016.01252
Fernandes, A. D., Reid, J. N., Macklaim, J. M., McMurrough, T. A., Edgell, D. R., and Gloor, G. B. (2014). Unifying the Analysis of High-Throughput Sequencing Datasets: Characterizing RNA-Seq, 16S rRNA Gene Sequencing and Selective Growth Experiments by Compositional Data Analysis. Microbiome 2, 15. doi:10.1186/2049-2618-2-15
Gihring, T. M., Zhang, G., Brandt, C. C., Brooks, S. C., Campbell, J. H., Carroll, S., et al. (2011). A Limited Microbial Consortium Is Responsible for Extended Bioreduction of Uranium in a Contaminated Aquifer. Appl. Environ. Microbiol. 77, 5955–5965. doi:10.1128/AEM.00220-11
Hassanshahian, M., Emtiazi, G., Caruso, G., and Cappello, S. (2014). Bioremediation (Bioaugmentation/biostimulation) Trials of Oil Polluted Seawater: A Mesocosm Simulation Study. Mar. Environ. Res. 95, 28–38. doi:10.1016/j.marenvres.2013.12.010
Hassenrück, C., Fink, A., Lichtschlag, A., Tegetmeyer, H. E., de Beer, D., Ramette, A., et al. (2016). Quantification of the Effects of Ocean Acidification on Sediment Microbial Communities in the Environment: the Importance of Ecosystem Approaches. FEMS Microbiol. Ecol. 92, fiw027. doi:10.1093/femsec/fiw027
Hemme, C. L., Tu, Q., Shi, Z., Qin, Y., Gao, W., Deng, Y., et al. (2015). Comparative Metagenomics Reveals Impact of Contaminants on Groundwater Microbiomes. Front. Microbiol. 6, 1205. doi:10.3389/fmicb.2015.01205
Herlemann, D. P. R., Geissinger, O., Ikeda-Ohtsubo, W., Kunin, V., Sun, H., Lapidus, A., et al. (2009). Genomic Analysis of "Elusimicrobium Minutum," the First Cultivated Representative of the Phylum "Elusimicrobia" (Formerly Termite Group 1). Aem 75, 2841–2849. doi:10.1128/AEM.02698-08
Hou, D., Huang, Z., Zeng, S., Liu, J., Wei, D., Deng, X., et al. (2017). Environmental Factors Shape Water Microbial Community Structure and Function in Shrimp Cultural Enclosure Ecosystems. Front. Microbiol. 8, 2359. doi:10.3389/fmicb.2017.02359
Idriss, H., Salih, I., and Sam, A. K. (2011). Study of Radon in Ground Water and Physicochemical Parameters in Khartoum State. J. Radioanal. Nucl. Chem. 290, 333–338. doi:10.1007/s10967-011-1295-4
Jiang, L., Song, M., Yang, L., Zhang, D., Sun, Y., Shen, Z., et al. (2016). Exploring the Influence of Environmental Factors on Bacterial Communities within the Rhizosphere of the Cu-Tolerant Plant, Elsholtzia Splendens. Sci. Rep. 6, 1–10. doi:10.1038/srep36302
Kalmbach, S., Manz, W., Wecke, J., and Szewzyk, U. (1999). Aquabacterium Gen. nov., with Description of Aquabacterium Citratiphilum Sp. nov., Aquabacterium Parvum Sp. Nov. And Aquabacterium Commune Sp. nov., Three In Situ Dominant Bacterial Species from the Berlin Drinking Water System. Int. J. Syst. Bacteriol. 49, 769–777. doi:10.1099/00207713-49-2-769
Kansal, S., Mehra, R., and Singh, N. P. (2012). Life Time Fatality Risk Assessment Due to Variation of Indoor Radon Concentration in Dwellings in Western Haryana, India. Appl. Radiat. Isot. 70, 1110–1112. doi:10.1016/j.apradiso.2011.11.057
Kitzinger, K., Koch, H., Lücker, S., Sedlacek, C. J., Herbold, C., Schwarz, J., et al. (2018). Characterization of the First "CandidatusNitrotoga" Isolate Reveals Metabolic Versatility and Separate Evolution of Widespread Nitrite-Oxidizing Bacteria. MBio 9. doi:10.1128/mBio.01186-18
Kolinko, S., Richter, M., Glöckner, F.-O., Brachmann, A., and Schüler, D. (2016). Single-cell Genomics of Uncultivated Deep-Branching Magnetotactic Bacteria Reveals a Conserved Set of Magnetosome Genes. Environ. Microbiol. 18, 21–37. doi:10.1111/1462-2920.12907
Li, J., Lin, S., and Qin, S. (2016). Characteristics of Sediment Bacterial Community in Response to Environmental Impacts in a Sewage Polluted River. J. Coastal Res. 74, 196–206. doi:10.2112/si74-017.1
Mahé, F., Rognes, T., Quince, C., de Vargas, C., and Dunthorn, M. (2014). Swarm: Robust and Fast Clustering Method for Amplicon-Based Studies. PeerJ 2, e593. doi:10.7717/peerj.593
Majumdar, R. K., Majumdar, N., and Mukherjee, A. L. (2000). Geoelectric Investigations in Bakreswar Geothermal Area, West Bengal, India. J. Appl. Geophys. 45, 187–202. doi:10.1016/S0926-9851(00)00028-8
Maleke, M., Williams, P., Castillo, J., Botes, E., Ojo, A., DeFlaun, M., et al. (2015). Optimization of a Bioremediation System of Soluble Uranium Based on the Biostimulation of an Indigenous Bacterial Community. Environ. Sci. Pollut. Res. 22, 8442–8450. doi:10.1007/s11356-014-3980-7
Naskar, A. K., Gazi, M., Barman, C., Chowdhury, S., Mondal, M., Ghosh, D., et al. (2018). Estimation of Underground Water Radon Danger in Bakreswar and Tantloi Geothermal Region, India. J. Radioanal. Nucl. Chem. 315, 273–283. doi:10.1007/s10967-017-5668-1
Nayak, T., De, D., Barman, C., Karmakar, P., Deb, A., and Dhal, P. K. (2020). Characterization of Indigenous Bacteria from Radon-Rich Groundwater and Their Tolerance to Physicochemical Stress. Int. J. Environ. Sci. Technol. 17, 1627–1636. doi:10.1007/s13762-019-02445-w
Negarestani, A., Setayeshi, S., Ghannadi-Maragheh, M., and Akashe, B. (2002). Layered Neural Networks Based Analysis of Radon Concentration and Environmental Parameters in Earthquake Prediction. J. Environ. Radioactivity 62, 225–233. doi:10.1016/S0265-931X(01)00165-5
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., et al. (2019). Package “Vegan” Title Community Ecology Package Version 2.5-6.
Poirel, L., Figueiredo, S., Cattoir, V., Carattoli, A., and Nordmann, P. (2008). Acinetobacter Radioresistens as a Silent Source of Carbapenem Resistance for Acinetobacter Spp. Antimicrob. Agents Chemother. 52, 1252–1256. doi:10.1128/AAC.01304-07
Pruesse, E., Peplies, J., and Glöckner, F. O. (2012). SINA: Accurate High-Throughput Multiple Sequence Alignment of Ribosomal RNA Genes. Bioinformatics 28, 1823–1829. doi:10.1093/bioinformatics/bts252
R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: https://www.R-project.org/ (Accessed September 28, 2017).
Rao, S., Chan, O. W., Lacap-Bugler, D. C., and Pointing, S. B. (2016). Radiation-Tolerant Bacteria Isolated from High Altitude Soil in Tibet. Indian J. Microbiol. 56, 508–512. doi:10.1007/s12088-016-0604-6
Reimann, C., Hall, G., Siewers, U., Bjorvatn, K., Morland, G., Skarphagen, H., et al. (1996). Radon, Fluoride and 62 Elements as Determined by ICP-MS in 145 Norwegian Hard Rock Groundwater Samples. Sci. Total Environ. 192, 1–19. doi:10.1016/0048-9697(96)05272-2
Rice, E. W., Baird, R. B., and Eaton, A. D. (2017). Standard Methods for the Examination of Water and Wastewater. 23rd Edition, United States: American public health association, American water works association, Water environmental federation.
Rocha, E. R., Selby, T., Coleman, J. P., and Smith, C. J. (1996). Oxidative Stress Response in an Anaerobe, Bacteroides Fragilis: A Role for Catalase in Protection against Hydrogen Peroxide. J. Bacteriol. 178, 6895–6903. doi:10.1128/jb.178.23.6895-6903.1996
RStudio Team (2017). RStudio: Integrated Development for R. RStudio, PBC, Boston, MA. Available at: http://www.rstudio.com/ (Accessed July 20, 2017).
Shukla, A., Parmar, P., and Saraf, M. (2017). Radiation, Radionuclides and Bacteria: An In-Perspective Review. J. Environ. Radioactivity 180, 27–35. doi:10.1016/j.jenvrad.2017.09.013
Shukla, M., Chaturvedi, R., Tamhane, D., Vyas, P., Archana, G., Apte, S., et al. (2007). Multiple-stress Tolerance of Ionizing Radiation-Resistant Bacterial Isolates Obtained from Various Habitats: Correlation between Stresses. Curr. Microbiol. 54, 142–148. doi:10.1007/s00284-006-0311-3
Singh, J., Singh, H., Singh, S., and Bajwa, B. S. (2008). Estimation of Uranium and Radon Concentration in Some Drinking Water Samples. Radiat. Measurements 43, S523–S526. doi:10.1016/j.radmeas.2008.04.004
Somlai, K., Tokonami, S., Ishikawa, T., Vancsura, P., Gáspár, M., Jobbágy, V., et al. (2007). 222Rn Concentrations of Water in the Balaton Highland and in the Southern Part of Hungary, and the Assessment of the Resulting Dose. Radiat. Measurements 42, 491–495. doi:10.1016/j.radmeas.2006.11.005
Song, J., Choo, Y.-J., and Cho, J.-C. (2008). Perlucidibaca Piscinae Gen. nov., Sp. nov., a Freshwater Bacterium Belonging to the Family Moraxellaceae. Int. J. Syst. Evol. Microbiol. 58, 97–102. doi:10.1099/ijs.0.65039-0
Thabayneh, K. M. (2015). Measurement of 222 Rn Concentration Levels in Drinking Water and the Associated Health Effects in the Southern Part of West Bank - Palestine. Appl. Radiat. Isot. 103, 48–53. doi:10.1016/j.apradiso.2015.05.007
Thompson, R. E. (2011). Epidemiological Evidence for Possible Radiation Hormesis from Radon Exposure: A Case-Control Study Conducted in Worcester, MA. Former. Nonlinearity Biol. 9, 59–75. doi:10.2203/dose-response.10-026.thompson
WHO (2011). Guidelines for Drinking-Water Quality. Chapter 9. Radiological Aspects, Geneva: WHO Press.
Wrighton, K. C., Thomas, B. C., Sharon, I., Miller, C. S., Castelle, C. J., VerBerkmoes, N. C., et al. (2012). Fermentation, Hydrogen, and Sulfur Metabolism in Multiple Uncultivated Bacterial Phyla. Science 337, 1661–1665. doi:10.1126/science.1224041
Wu, M. L., Van Teeseling, M. C. F., Willems, M. J. R., Van Donselaar, E. G., Klingl, A., Rachel, R., et al. (2012). Ultrastructure of the Denitrifying Methanotroph "Candidatus Methylomirabilis Oxyfera," a Novel Polygon-Shaped Bacterium. J. Bacteriol. 194, 284–291. doi:10.1128/JB.05816-11
Yan, T., Fields, M. W., Wu, L., Zu, Y., Tiedje, J. M., and Zhou, J. (2003). Molecular Diversity and Characterization of Nitrite Reductase Gene Fragments (nirK and nirS) from Nitrate- and Uranium-Contaminated Groundwater. Environ. Microbiol. 5, 13–24. doi:10.1046/j.1462-2920.2003.00393.x
Yan, X., Luo, X., and Zhao, M. (2016). Metagenomic Analysis of Microbial Community in Uranium-Contaminated Soil. Appl. Microbiol. Biotechnol. 100, 299–310. doi:10.1007/s00253-015-7003-5
Keywords: radon contamination, drinking water, heavy metal, bioremediation, microbial community, 16S rRNA gene amplicon sequencing
Citation: Nayak T, De D, Karmakar P, Deb A and Dhal PK (2021) Microbial Communities of the Drinking Water With Gradient Radon Concentration Are Primarily Contributed by Radon and Heavy Metal Content. Front. Environ. Sci. 9:576400. doi: 10.3389/fenvs.2021.576400
Received: 26 June 2020; Accepted: 26 April 2021;
Published: 17 May 2021.
Edited by:
Andrew Hursthouse, University of the West of Scotland, United KingdomReviewed by:
Zhi Wang, Innovation Academy for Precision Measurement Science and Technology (CAS), ChinaAvishek Dutta, University of California, San Diego, United States
Copyright © 2021 Nayak, De, Karmakar, Deb and Dhal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paltu Kumar Dhal, paltuk.dhal@jadavpuruniversity.in
 Tilak Nayak
Tilak Nayak Debjit De1
Debjit De1  Paltu Kumar Dhal
Paltu Kumar Dhal