Abstract
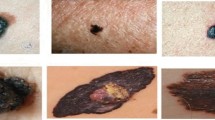
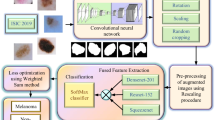
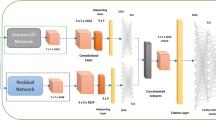
Skin lesion is one of the severe diseases which in many cases endanger the lives of patients on a worldwide extent. Early detection of disease in dermoscopy images can significantly increase the survival rate. However, the accurate detection of disease is highly challenging due to the following reasons: e.g., visual similarity between different classes of disease (e.g., melanoma and non-melanoma lesions), low contrast between lesions and skin, background noise, and artifacts. Machine learning models based on convolutional neural networks (CNN) have been widely used for automatic recognition of lesion diseases with high accuracy in comparison to conventional machine learning methods. In this research, we proposed a new preprocessing technique in order to extract the region of interest (RoI) of skin lesion dataset. We compare the performance of the most state-of-the-art CNN classifiers with two datasets which contain (1) raw, and (2) RoI extracted images. Our experiment results show that training CNN models by RoI extracted dataset can improve the accuracy of the prediction (e.g., InceptionResNetV2, 2.18% improvement). Moreover, it significantly decreases the evaluation (inference) and training time of classifiers as well.





Similar content being viewed by others
References
Zeinali B, Ayatollahi A, Kakooei M (2014) A novel method of applying directional filter bank (dfb) for finger-knuckle-print (fkp) recognition. In: 2014 22nd Iranian conference on electrical engineering (ICEE), pp 500–504
Codella N, Rotemberg V, Tschandl P, Celebi ME, Dusza S, Gutman D, Helba B, Kalloo A, Liopyris K, Marchetti M et al (2019) Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv:1902.03368
Gutman D, Codella NC, Celebi E, Helba B, Marchetti M, Mishra N, Halpern A (2016) Skin lesion analysis toward melanoma detection: a challenge at the international symposium on biomedical imaging (isbi) 2016, hosted by the international skin imaging collaboration (isic). arXiv:1605.01397
Tschandl P, Rosendahl C, Kittler H (2018) The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data 5:180161
Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H et al (2018) Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), IEEE, pp 168–172
Combalia M, Codella NC, Rotemberg V, Helba B, Vilaplana V, Reiter O, Halpern AC, Puig S, Malvehy J (2019) Bcn20000: dermoscopic lesions in the wild. arXiv:1908.02288
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2017) Brain tumor segmentation and radiomics survival prediction: contribution to the brats 2017 challenge. In: International MICCAI brainlesion workshop. Springer, pp 287–297
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2017) Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848
Ng H, Ong S, Foong K, Goh P, Nowinski W (2006) Medical image segmentation using k-means clustering and improved watershed algorithm. In: 2006 IEEE southwest symposium on image analysis and interpretation, IEEE, pp 61–65
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 234–241
Yu L, Chen H, Dou Q, Qin J, Heng P. -A. (2016) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imag 36(4):994–1004
Bi L, Kim J, Ahn E, Kumar A, Fulham M, Feng D (2017) Dermoscopic image segmentation via multistage fully convolutional networks. IEEE Trans Biomed Eng 64(9):2065–2074
Yuan Y, Chao M, Lo Y-C (2017) Automatic skin lesion segmentation using deep fully convolutional networks with jaccard distance. IEEE Trans Med Imaging 36(9):1876–1886
Goyal M, Yap MH, Hassanpour S (2017) Multi-class semantic segmentation of skin lesions via fully convolutional networks. arXiv:1711.10449
Vesal S, Patil SM, Ravikumar N, Maier AK (2018) A multi-task framework for skin lesion detection and segmentation. In: OR 2.0 Context-aware operating theaters, computer assisted robotic endoscopy, clinical image-based procedures, and skin image analysis, Springer, pp 285–293
Soudani A, Barhoumi W (2019) An image-based segmentation recommender using crowdsourcing and transfer learning for skin lesion extraction. Expert Syst Appl 118:400–410
Zhang J, Hu J (2008) Image segmentation based on 2d otsu method with histogram analysis. In: 2008 International conference on computer science and software engineering, vol 6. IEEE, pp 105–108
Haggerty JM, Wang XN, Dickinson A, O’Malley CJ, Martin EB (2014) Segmentation of epidermal tissue with histopathological damage in images of haematoxylin and eosin stained human skin. BMC Med Imaging 14(1):7
Bindu CH, Prasad KS (2012) An efficient medical image segmentation using conventional otsu method. Int J Adv Sci Technol 38(1):67–74
Premaladha J, Ravichandran K (2016) Novel approaches for diagnosing melanoma skin lesions through supervised and deep learning algorithms. J Med Syst 40(4):96
Buza E, Akagic A, Omanovic S (2017) Skin detection based on image color segmentation with histogram and k-means clustering. In: 2017 10th International conference on electrical and electronics engineering (ELECO), pp 1181–1186
McGuinness K, O’Connor NE (2010) A comparative evaluation of interactive segmentation algorithms. Pattern Recogn 43(2):434–444. interactive Imaging and Vision. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S0031320309000818
Berseth M (2017) Isic 2017-skin lesion analysis towards melanoma detection. arXiv:1703.00523
Zhao T, Gao D, Wang J, Tin Z (2018) Lung segmentation in ct images using a fully convolutional neural network with multi-instance and conditional adversary loss. In: 2018 IEEE 15th International symposium on biomedical imaging (ISBI 2018), IEEE, pp 505–509
Xiao X, Lian S, Luo Z, Li S (2018) Weighted res-unet for high-quality retina vessel segmentation. In: 2018 9th International conference on information technology in medicine and education (ITME), IEEE, pp 327–331
Li X, Chen H, Qi X, Dou Q, Fu C-W, Heng P-A (2018) H-denseunet: hybrid densely connected unet for liver and tumor segmentation from ct volumes. IEEE Trans Med Imaging 37(12):2663–2674
Ibtehaz N, Rahman MS (2020) Multiresunet: Rethinking the u-net architecture for multimodal biomedical image segmentation. Neural Netw 121:74–87
Jafari MH, Karimi N, Nasr-Esfahani E, Samavi S, Soroushmehr SMR, Ward K, Najarian K (2016) Skin lesion segmentation in clinical images using deep learning. In: 2016 23rd International conference on pattern recognition (ICPR), IEEE, pp 337–342
Kawahara J, Hamarneh G (2016) Multi-resolution-tract cnn with hybrid pretrained and skin-lesion trained layers. In: International workshop on machine learning in medical imaging, Springer, pp 164–171
Saba T, Khan MA, Rehman A, Marie-Sainte SL (2019) Region extraction and classification of skin cancer: a heterogeneous framework of deep cnn features fusion and reduction. J Med Syst 43(9):289
Mahbod A, Schaefer G, Wang C, Ecker R, Dorffner G, Ellinger I (2020) Investigating and exploiting image resolution for transfer learning-based skin lesion classification
Hosny KM, Kassem MA, Foaud MM (2018) Skin cancer classification using deep learning and transfer learning. In: 2018 9th Cairo international biomedical engineering conference (CIBEC), pp 90–93
Adegun AA, Viriri S (2020) Deep learning-based system for automatic melanoma detection. IEEE Access 8:7160–7172
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: NIPS
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence
Tan M, Le QV (2019) Efficientnet: rethinking model scaling for convolutional neural networks. arXiv:1905.11946
Chollet F (2016) Xception: Deep learning with depthwise separable convolutions. arXiv:1610.02357
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2015) Rethinking the inception architecture for computer vision. arXiv:1512.00567
Zoph B, Vasudevan V, Shlens J, Le QV (2017) Learning transferable architectures for scalable image recognition. arXiv:1707.07012
Huang G, Liu Z, van der Maaten L, Weinberger KQ (2016) Densely connected convolutional networks. arXiv:1608.06993
He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition. arXiv:1512.03385
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Chollet F, et al. (2015) Keras. https://keras.io, [accessed April 1 2020]
Liu Y, Yao X (1999) Ensemble learning via negative correlation. Neural Netw 12(10):1399–1404
Acknowledgements
Effort sponsored in whole or in part by United States Special Operations Command (USSOCOM), under Partnership Intermediary Agreement No. H92222-15-3-0001-01. The U.S. Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation thereon.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The authors used the data publicly available for their study and did not collect data from any human participant or animal.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Disclaimer
The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the United States Special Operations Command
Rights and permissions
About this article
Cite this article
Zanddizari, H., Nguyen, N., Zeinali, B. et al. A new preprocessing approach to improve the performance of CNN-based skin lesion classification. Med Biol Eng Comput 59, 1123–1131 (2021). https://doi.org/10.1007/s11517-021-02355-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-021-02355-5