Abstract
The iron dependence of antibiotic-resistant microbes represents an Achilles’ heel that can be exploited broadly. The growing global problem of antibiotic resistance of microbial pathogens wherein microbes become resistant to the very antibiotics used against them during infection is linked not only to our health uses but also to agribusiness practices and the changing environment. Here we review mechanisms of microbial iron acquisition and host iron withdrawal defense, and the influence of iron withdrawal on the antimicrobial activity of antibiotics. Antibiotic-resistant microbes are unaltered in their iron requirements, but iron withdrawal from microbes enhances the activities of various antibiotics and importantly suppresses outgrowth of antibiotic-exposed resistant microbial survivors. Of the three therapeutic approaches available to exploit microbial iron susceptibility, including (1) use of gallium as a non-functional iron analogue, (2) Trojan horse conjugates of microbial siderophores carrying antibiotics, and (3) new generation iron chelators, purposely designed as anti-microbials, the latter offers various advantages. For instance, these novel anti-microbial chelators overcome the limitations of conventional clinically-used hematological chelators which display host toxicity and are not useful antimicrobials. 3-Hydroxypyridin-4-one-containing polymeric chelators appear to have the highest potential. DIBI (developmental code name) is a well-developed lead candidate, being a low molecular weight, water-soluble copolymer with enhanced iron binding characteristics, strong anti-microbial and anti-inflammatory activities, low toxicity for animals and demonstrated freedom from microbial resistance development. DIBI has been shown to enhance antibiotic efficacy for antibiotic-resistant microbes during infection, and it also prevents recovery growth and resistance development during microbe exposure to various antibiotics. Because DIBI bolsters innate iron withdrawal defenses of the infected host, it has potential to provide a host-directed anti-infective therapy.
Similar content being viewed by others
Introduction
Antibiotic resistance of microbial pathogens wherein a microbe becomes resistant to the antimicrobial activity of an antibiotic used against it during infection is a growing global problem with morbidity and mortality rapidly increasing as effective antibiotic choices dwindle (Barrasa-Villar et al. 2017; Friedman et al. 2016). In perspective, our antibiotic resistance problem is not surprising given most antibiotics are, or are derived from, natural microbial products (Katz and Baltz 2016; Movassaghi and van der Donk 2018). Antibiotics are also employed in a microbe-targeted approach where the antibiotic targets one or only a few closely related microbial target(s) (Luepke et al. 2017). From this relatively narrow microbe-targeted approach, positive selection pressure from the antibiotic employed favors survival and outgrowth of antibiotic-resistant clones (Bengtsson-Palme et al. 2018).
The problems of antibiotic resistance are also inextricably linked to human and animal health antibiotic use/misuse, agribusiness use practices, and the environment in general. Many pathogens are opportunistic and otherwise commensal microbes found naturally in soil and water habitats. Resistance can arise from use of sub-inhibitory concentrations of antibiotics used as growth promoters in animal husbandry or from antibiotic residues released to the environment in domestic or livestock sewage, etc. (Manyi-Loh et al. 2018). Furthermore, climate change is driving resistance spread as higher surface water temperatures favor increased growth and spread of genetic resistance attributes among, for example, Vibrio spp. including V. cholerae that cause serious water and food-borne, sometimes pandemic, enteric infections (Baker-Austin et al. 2018).
Given these various problems, new alternative approaches to conventional antibiotics are urgently needed to address resistance. Of various possibilities, the top ranked alternatives including use of bacteriophages, anti-bacterial peptides, and lysins have yet to yield useful new therapeutics despite over 10 years of investigation (Czaplewski et al. 2016). It is interesting to note that these are also relatively narrow microbe-targeted approaches.
A more recent alternative approach is to target pathogens more broadly based on their irreplaceable and heightened requirements for a growth essential trace metal during infection (Holbein et al. 2021). Iron requirements of pathogens stand out as the top candidate, given almost all bacterial and fungal pathogens have irreplaceable iron requirements for their growth and survival (Ganz 2018; Weinberg 2009). Iron needs for a multitude of essential iron-dependent targets (mostly enzymes) in microbes are evident during infection, and this iron must be obtained from the infected host (Ganz 2018; Palmer and Skaar 2016; Weinberg 1978, 2009). Importantly, vertebrate hosts deploy innate iron withdrawal defense mechanisms that attempt to limit infection (Ganz and Nemeth 2015; Holbein 1980; Letendre and Holbein 1984; Nairz et al. 2018). The absolute iron dependency of microbes on the one hand and the iron withdrawal defenses of the host on the other hand provide the basis for a new anti-infective approach that is not solely dependent on microbe targeting but rather introduces a host-directed approach through bolstering natural defenses.
There are three separate therapeutic approaches that can exploit the iron dependence of antibiotic-resistant (or sensitive) microbes: (1) gallium a non-functional iron analogue metal, as administered and taken up by but not useful to microbes; (2) synthetic copies of microbial iron acquisition siderophores, chemically linked to antibiotics resulting in new Trojan horse carriers of antibiotics, and (3) new generation purpose-designed anti-microbial iron chelating compounds.
The clinical iron chelators in current use for other medical needs (deferoxamine, DFO; deferiprone, DFP, and deferasirox, DEF) have been investigated for repurposing as anti-infectives with poor results. These chelators, not being first developed as anti-infectives, are only weakly anti-infective or worse, actually utilized for iron by some microbes. These also present toxicity concerns as they do not preferentially access the dynamic extracellular host iron pools which are most relevant to infection but rather inappropriately access host intracellular iron pools leading to their toxicity.
For utility as anti-infectives, new iron chelators need to preferentially target and withdraw microbial accessible iron pools but not interfere with longer-term host iron homeostasis. In this regard, new generation iron chelators utilizing hydroxypyridinone metal binding groups have been purpose-designed as anti-infectives. Of these, the hydroxypyridinone-functionalized polymers have demonstrated particular promise. DIBI (developmental code name) is a lead example that has been extensively developed and tested in a variety of animal infection models. DIBI preferentially binds extracellular iron and much more strongly than conventional chelators, its bound iron is inaccessible to microbes making it 1000X more antimicrobial than current choices, and it has been shown to be non-toxic when administered systemically to animals.
Here, we review antibiotic resistance of microbial pathogens in the context of their iron acquisition and host iron withdrawal defenses. Of major relevance is the iron requirements of antibiotic-resistant microbes are like their antibiotic sensitive counterparts and iron restriction to microbes does not impede but rather enhances antibiotic activity including with antibiotic-resistant isolates. Very important in this context is that iron withdrawal by chelators during antibiotic exposure to microbes restricts microbial recovery growth thus also suppressing development of antibiotic resistance. Thus, iron dependency of antibiotic-resistant microbes represents their Achilles’ heel to exploit therapeutically.
New generation antimicrobial iron chelators such as DIBI have high promise to provide new host-directed therapies for infection. Host-directed therapies, i.e., those that work with and enhance innate host defense mechanisms, offer broader and longer-term potential, i.e., as opposed to continued reliance on narrow-target microbe-directed antibiotic therapies. This article is an abridged and updated version of our recent more comprehensive review (Holbein et al. 2021).
Microbial iron acquisition and host iron withdrawal during infection
It is quite probable that spatial and temporal fluctuations of iron availability in a single host organism do occur and that these are sufficient to permit microbial growth in one portion and microbial death in another part of the same tissue
E.D. Weinberg (1978).
In the now over 40 years following Eugene Weinberg’s landmark review on iron in infection (Weinberg 1978), much has been learned as to the mechanisms that various microbial pathogens deploy to obtain iron from their infected host and how the host in response deploys iron withholding strategies to limit infection. Weinberg’s original hypothesis (quoted above) has proved especially insightful as we now better understand that different iron pools within the body can play quite differing and pivotal roles. For example, a relatively small reactive intracellular iron pool plays a key microbiocidal role through iron-driven Reactive Oxygen Species (ROS) generation in host defensive phagocytes; these reactive oxygen species kill the phagocytosed bacteria (Cronin et al. 2019). At the same time, other extracellular iron pools of the host can be accessed by microbes for their growth. Microbes deploy various acquisition mechanisms to obtain this extracellular iron, as most microbial pathogens grow extracellularly either exclusively or at least during their initial stages of infection.
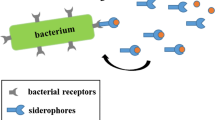
Bacteria have evolved various strategies and mechanisms to obtain needed iron from their host. These include: (1) siderophore production and uptake, (2) iron acquisition from heme, (3) iron acquisition from host iron sequestering proteins such as lactoferrin and transferrin, and (4) reduction of ferric to ferrous iron (the ferrous form being more soluble/transportable).
Bacterial siderophores
Based on their chemical functionality as to iron binding groups, siderophores can be divided into three general chemical classes: hydroxamates, catecholates, and hydroxycarbonates (Bilitewski et al. 2017; Caza and Kronstad 2013; Foley and Simeonov 2012) as shown in Fig. 1. More detailed reviews on the range of siderophores produced by different microbes can be found elsewhere (Ellermann and Arthur 2017; Holbein et al. 2021; Holden and Bachman 2015; Johnstone and Nolan 2015; Wilson et al. 2016).
Siderophores typically chelate ferric iron (Fe(III)) with high-affinity (see Fig. 1) and therefore often out-compete the iron sequestering molecules of the host (Ellermann and Arthur 2017; Wilson et al. 2016). After binding ferric iron, ferri-siderophores are retrieved from the extracellular environment by their cognate receptors either on the outer membrane of Gram-negative bacteria or on cell wall-associated receptors of Gram-positive bacteria. These receptors facilitate translocation of the iron complex into the periplasmic space (Bairwa et al. 2017; Caza and Kronstad 2013; Foley and Simeonov 2012; Holden and Bachman 2015; Johnstone and Nolan 2015; Khan et al. 2018; Sheldon and Heinrichs 2015; Wilson et al. 2016). The specific Gram-negative bacterial surface receptors (such as FepA, IroN, and PfeA from Escherichia coli, Salmonella enterica and Pseudomonas aeruginosa, respectively) are all members of the well-known TonB-dependent transporter family and are usually regulated by the ferric uptake regulator protein, Fur (Caza and Kronstad 2013).
Periplasmic ferri-siderophore binding proteins then shuttle the molecules to specific cell membrane-associated ABC transporters, which hydrolyse ATP to drive transport of the iron bearing cargo into the bacterial cytosol (Foley and Simeonov 2012). Once the molecule reaches the intracellular space, its iron can be released by physical degradation of the siderophore or through chemical reduction of iron(III) to iron(II) using ferri-siderophore reductases or esterases (Caza and Kronstad 2013; Foley and Simeonov 2012; Kelson et al. 2013; Zhou et al. 2012).
Given the importance of iron, microbial pathogens often deploy multiple iron acquisition systems or siderophores to ensure iron acquisition from the infected host (Bairwa et al. 2017; Caza and Kronstad 2013; Sheldon and Heinrichs 2015; Sheldon et al. 2016). While different microbial receptors are often selective for a particular ferri-siderophore complexes, some bacteria possess broader receptor/uptake systems for various siderophores secreted by other bacteria (i.e., xenosiderophores) to achieve what has been termed iron piracy (Caza and Kronstad 2013). In addition, some bacteria release siderophores to deprive microbial competitors of iron availability in the community, for example in the soil.
Some microbes such as Listeria monocytogenes and Candida albicans do not produce siderophores; instead utilizing heterogeneous xenosiderophores produced by other microbes (Caza and Kronstad 2013). Alternatively, Neisseria meningitidis does not produce a siderophore, but it can directly access iron on host transferrin (Holbein 1981). Yet other microbial pathogens such as P. aeruginosa although producing its own siderophores can also use xenosiderophores to better compete for iron (Caza and Kronstad 2013). Vibrio cholerae has been shown to utilize the TonB-dependent transport system for vibriobactin, its endogenous siderophore as well as several siderophores produced by other microorganisms (Wyckoff et al. 2007), and this so-called siderophore piracy enhances its ability to grow in iron-limiting culture and to colonize mice in the presence of siderophore-producing Escherichia coli. This ability might enhance the ability of V. cholerae to grow in sewage contaminated water as associated with cholera outbreaks.
Bacterial heme utilization
Most of our body iron is incorporated into biological molecules such as hemoglobin with a porphyrin ring as ferriprotoporphyrin IX (heme) and this is not directly accessible by microbes (Kelson et al. 2013). Heme iron is accessible by intracellular pathogens or by extracellular pathogens that produce cytotoxins (such as cytolysins or hemolysins) that lyse host cells (e.g., red blood cells) to release hemeproteins. Implicitly lysin production would require bacterial growth and thus iron for the initial infection and that phase of infection would likely be dependent on non-heme host iron supplies. In severe sepsis, free heme levels in plasma can also be elevated due to tissue damage (Larsen et al. 2010), and therefore, under these conditions additional extracellular heme would presumably become more available to extracellular pathogens.
Bacteria have developed various strategies to obtain heme iron from their host during infection by targeting heme, hemoglobin, or complexes containing these molecules (e.g., haptoglobin–hemoglobin, hemopexin–heme) as reviewed previously (Holbein et al. 2021).
It has been suggested that S. aureus and some strains of A. baumannii may have more active iron acquisition systems using heme-based versus siderophore-based acquisition (de Leseleuc et al. 2014; Kelson et al. 2013). V. cholerae that utilizes various siderophores also utilizes heme as an iron source (Wyckoff et al. 2007). The body does possess defenses to heme access with hemopexin, a liver protein released during inflammation which binds to free heme to eliminate it from systemic circulation thus preventing oxidative stress from release of heme bound iron (Tolosano and Altruda 2002). Addition of exogenous hemopexin has been shown to modulate experimental P. aeruginosa pneumonia in mice (Wagener et al. 2018).
Microbial utilization of transferrins and other host iron sources
Various bacterial pathogens can utilize non-heme, iron-containing proteins as sources of iron, such as the transferrin class iron carrying proteins (transferrin in blood plasma and lactoferrin in exocrine secretions) as well as intracellular ferritins (Caza and Kronstad 2013; Chitambar 2010; Holbein 1981). Acquisition from transferrin and lactoferrin is typically through receptors for these proteins utilizing mechanisms as reviewed recently (Holbein et al. 2021).
Bacteria can also elaborate cell surface ferric reductases to reduce free ferric iron to ferrous iron which can then be taken up more readily and utilized by the bacterial cell (Kelson et al. 2013; Sheldon et al. 2016). Ferrous iron ions are believed to diffuse freely through the outer membrane of Gram-negative bacteria, with subsequent transport through the inner membrane by the ABC transporter FeoABC (Caza and Kronstad 2013) which is under control of fnr (for anaerobes) and fur regulatory elements (Caza and Kronstad 2013).
In common with bacteria, pathogenic fungi possess a number of mechanisms to acquire iron from different sources in the mammalian host with many of these mechanisms sharing functional similarities with bacterial pathogens (Caza and Kronstad 2013; Holbein et al. 2021).
Host iron withdrawal during infection
Invasion of vertebrate animals by microbes triggers a host inflammatory response with an associated hypoferremic iron withdrawal, whereby extracellular iron levels, particularly those in the circulating transferrin pool, are lowered to limit microbial access to iron. The ability of the hypoferremic response to directly control infection was first demonstrated with N. meningitidis, the Gram-negative obligate bacterial parasite that causes human cerebrospinal meningitis and sepsis. A hypervirulent clinical isolate of N. meningitidis produced a rapid bacteremia in mice that peaked at 6–9 h, and then, this infection completely resolved by 18 h while circulating blood transferrin iron levels (Fe-Tf) dropped to near undetectable levels in concert with disappearance of the infection (Holbein 1980). Furthermore, addition of either exogenous Fe-dextran (Holbein 1980) or Fe-transferrin but not iron-free Apo-transferrin (Holbein 1981) supported continued infection resulting in fulminant sepsis and fatality. Investigation of the mechanism of this hypoferremic response during meningococcal infection using radiolabeled iron demonstrated that the drop in the Fe-transferrin pool was not due to an accelerated removal of this iron pool by the host, but rather it was associated with an impaired delivery/resupply of new iron to this dynamic transferrin pool from intracellular sources (Letendre and Holbein 1984); this Fe-transferrin pool has a high iron turnover rate, i.e., T1/2 for iron of only 0.7 h (Letendre and Holbein 1983).
Some years later, the vertebrate host hypoferremic response was shown to be part of the more generalized inflammatory responses trigged by infection or by bacterial cell components. In addition to actual infection, both lipopolysaccharide (LPS) from Gram-negative bacteria and lipoteichoic acid (LTA) from Gram-positive bacteria induce leukocyte activation and adhesion to blood vessels and also stimulate production of inflammatory cytokines including interlukin-6 (IL-6) in mice in the absence of viable bacteria (Fokam and Hoskin 2020). Lipopolysaccharide binds to the macrophage toll-like receptor 4 (TLR4). TLR4 activation then causes translocation of nuclear factor-kB (NF-kB) into the nucleus of innate immune cells and transcription, translation, and cellular release of inflammatory mediators such as tumor necrosis factor-alpha (TNF-α) and interlukin-6. Interlukin-6 is known to upregulate hepcidin synthesis in the liver (Johnson and Wessling-Resnick 2012). Hepcidin is a host hepatocyte produced antimicrobial peptide and also the main regulator of vertebrate host iron hemostasis. Increased hepcidin levels then cause cellular internalization and lysosomal degradation of ferroportin, the cell membrane-associated transporter protein responsible for iron release from intracellular to extracellular (transferrin) compartments. This results in lowering of transferrin iron levels, i.e., the hypoferremic response. Macrophages are now known to play important central control roles in these inflammatory responses as well for overall iron homeostasis (Sukhbaatar and Weichhart 2018). A key overall result from activation of these pathways is a decreased extracellular iron availability (Barton and Acton 2019).
Importantly, this host hypoferremic response occurs very early in infection while microbes are in battle to obtain their initial iron supplies from host iron pools. In this regard, it is also important to appreciate that most microbes do not typically store excess iron for future needs and generally arise from iron-limited environments as they enter the body to infect. Thus, when microbes find the opportunity to initiate infection on or within a vertebrate host, they require immediate access to host iron, i.e., to grow and evade host defense clearance mechanisms. On this basis, iron access by microbes and host iron withdrawal defenses are very important during the early (including subclinical) stages of infection.
Other factors involved in limiting extracellular iron availability include lactoferrin and lipocalin (siderocalin). Constitutively high levels of lactoferrin are found in most mucosal surface secretions, including tears, saliva, bile, and breast milk (Ward et al. 2002). Pro-inflammatory cytokines such as TNF-α also induce the release of lactoferrin from neutrophilic granules at the site of infection (Afeltra et al. 1997). Accordingly, the addition of exogenous lactoferrin has been shown to reduce pulmonary M. tuberculosis burden in a mouse model of iron overload (Schaible et al. 2002) and growth of S. aureus in the mammary system of cows with mastitis (Lacasse et al. 2008). The capacity of lactoferrin to sequester iron also has been implicated in the ability of patients with cystic fibrosis to control opportunistic P. aeruginosa infection and biofilm formation in the respiratory tract (Britigan et al. 1993).
Iron in systemic inflammation and sepsis
Sepsis is defined as life-threatening organ dysfunction caused by a dysregulated host response to infection (Singer et al. 2016) and considered the “worst case” of an infection, where the body’s overwhelming immune response leads to tissue damage, organ failure, and death. With over 1.5 million sepsis hospital stays per year, sepsis costs total $27 billion each year in the USA alone (Paoli et al. 2018). At present, there is no FDA-approved therapeutic for sepsis available.
There is increasing clinical evidence that host iron dysregulation plays important roles in susceptibility to sepsis and its disease course (Brandtner et al. 2020). Iron plays Janus-faced roles in sepsis. Physiological intracellular iron levels are beneficial for the efficient production of bactericidal reactive oxygen species utilizing the Haber–Weiss and Fenton reactions with iron as the catalytic co-factor (Fokam and Hoskin 2020). Nonetheless, reactive oxygen species over-production, a common event in sepsis due to the dysregulation of the immune response can harm healthy tissues and contribute to organ dysfunction and multiorgan failure (Dickson and Zhou 2020). Therefore, iron restriction is considered a potential target in sepsis therapy (Scott et al. 2020). As outlined above, the approach of iron withdrawal by chelators could also reduce bacterial growth which is the main goal of sepsis therapy. Limitations of this approach are the hypo-inflammatory phase of sepsis (immuno-paralysis) and most likely in conditions with severe true anemia.
Clinical findings have confirmed that the known physiological mechanisms of iron restriction in infection also have implications for sepsis as critically ill sepsis patients generally have low systemic iron levels as consequence of the host defense to infection (Tacke et al. 2016). This physiological response to infection is often misunderstood and patients often receive (too) early treatment with blood transfusions or even erythropoietin for clinical iron deficiency, i.e., anemia. Not surprisingly, these therapies did not improve the outcome of sepsis in randomized controlled studies (Jandu et al. 2018; Pearl 2014).
Animal experiments have shown beneficial effects of iron chelation in models of sepsis with reduced hyper-inflammation and improved survival confirming the concept of the dual role of iron deprivation in systemic inflammation/infection: reduction in bacterial growth and attenuation of reactive oxygen species overproduction. Specifically, studies from our group using the novel iron chelator, DIBI, which binds iron in a form inaccessible for bacteria, showed a significant reduction in leukocyte over-activation and excess systemic cytokine levels in models of both endotoxin-induced and surgical sepsis (Islam et al. 2016; Thorburn et al. 2017). More recently, we have shown that iron chelation by DIBI is beneficial in sepsis induced by several other bacterial toxins (Fokam et al. 2020b) and improves survival in preclinical sepsis models (Fokam et al. 2020a). These findings were confirmed in experiments with DIBI compared to the classical iron chelators, deferiprone, deferasirox and deferoxamine (Lehmann et al. 2021). In these studies, DIBI’s superior iron binding characteristics, its low host toxicity and its higher antimicrobial activities as compared to these conventional clinical chelators (see Table 1) were consistent with the findings of reduced bacterial burden and better modulation of the excess inflammatory responses in experimental polymicrobial sepsis.
Clinical trials with iron chelators to treat sepsis have not yet been carried out but these now appear warranted. The body of evidence for the role of iron in sepsis is growing. However, despite this increased knowledge, iron levels and related clinical markers of iron metabolism are not yet routinely measured in sepsis patients and such measurements would also appear to be warranted. In particular, patients with underlying diseases are at increased risk of sepsis-related complications: dysregulated iron metabolism with elevated plasma and/or increased intracellular iron stores is a feature of both, diabetes (Altamura et al. 2017) and cardiovascular disease (Wunderer et al. 2020), and has also been implicated in hypertension (Sangartit et al. 2016).
Exploiting essential microbial iron needs
We have previously discussed the iron requirements of microbial pathogens in detail (Holbein et al. 2021). Pathogenic microbes are completely dependent on iron acquisition for their virulence and growth during infection, and this essential requirement representing a microbial Achilles’ heel can be targeted in three separate ways.
Gallium iron mimic
Iron(III) can be displaced by gallium(III) (Ga) in most iron complexes, but gallium does not have the essential redox properties of iron(III), and thus, gallium-substituted complexes are non-functional. Gallium(III) salts or gallium organo-complexes have been investigated as anti-infectives (Banin et al. 2008; Chitambar 2016, 2017; Kelson et al. 2013; Rangel-Vega et al. 2015; Stojiljkovic et al. 1999). We have reviewed these investigations more fully previously (Holbein et al. 2021).
Despite some promising results, there are several challenges to using gallium as an alternative to antibiotics. Not all bacterial strains are susceptible to gallium, for example, some A. baumannii strains can circumvent gallium nitrate activity with a heme receptor (de Leseleuc et al. 2014). In addition, there is the toxicity concern when high or prolonged doses of gallium might be used since the toxicity of gallium and its compounds have yet to be fully studied (Chitambar 2016, 2017). Gallium would presumably also displace host iron, and since there are no excretion mechanisms available for iron, gallium parenterally administered could accumulate in the body leading to toxicity. Further elucidation of the uptake and metabolism mechanisms of iron and gallium in bacteria will help define and optimize potential use of gallium complexes in vivo. Additional evaluation of gallium-based drugs, particularly as related to potential host toxicity issues and their effects on innate host iron withdrawal defense mechanisms, is needed to establish whether potential gallium-based therapeutics have a significant place in the treatment of infections in humans.
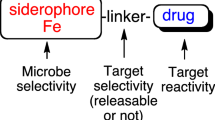
Trojan horse siderophore–antibiotic conjugates
Siderophore–antibiotic conjugates utilize a siderophore structure essentially as a bait and vehicle to deliver an antibiotic cargo into the periplasm or cytosol of bacterial cells through the cognate cell surface siderophore-specific receptors, akin to a “Trojan horse” (Budzikiewicz 2001; Mollmann et al. 2009). A range of natural and synthetic siderophore–antibiotic conjugates have been identified or synthesized over the past 50 years (Johnstone and Nolan 2015). Because of their potential attractiveness, rationally designed synthetic siderophore–antibiotic conjugates comprising different classes of antibiotics and relevant or specific siderophores of targeted pathogen have been constructed. We have previously reviewed various siderophore–antibiotic candidates (Holbein et al. 2021). To date, siderophore–lactam conjugates have been the most investigated and developed. Since our previous review (Holbein et al. 2021), four compounds, cefetecol, BAL30072, GSK3342830, and cefiderocol (Powers 2020), have reached clinical trials. Cefiderocol (Fetroja®) is the first FDA-approved novel, synthetic siderophore–antibiotic conjugate. Its cephalosporin moiety inhibits bacterial cell wall synthesis by binding to penicillin-binding proteins (PBP) and inhibiting peptidoglycan synthesis, whereas the siderophore (catechol) moiety facilitates the entry into bacterial cells through active iron transporters in E. coli and P. aeruginosa (Bonomo 2019; Naseer et al. 2020).
Despite these promising results, selection of the appropriate siderophore vehicle remains challenging due to: (1) the rapid emergence of potential resistance due to the presence of multiple siderophore receptors in bacteria; (2) the existence of bacterial “cheaters” which take siderophores from other bacteria rather than their own, a common feature in polymicrobial infections; and (3) the potential down-regulation of the siderophore receptors on bacterial cell surface (Budzikiewicz 2001; Foley and Simeonov 2012). While new siderophore–antibiotic conjugates have potential to deliver antibiotics to susceptible microbes, the likelihood for resistance development can be expected to be similar to what has already been experienced from other conventional antibiotics.
Iron chelators to intercept and deny iron to microbes
Natural microbial siderophores such as deferoxamine (DFO) and the synthetic chelators, deferiprone (DFP), and deferasirox (DEF) have been clinically approved and are currently used for the treatment of various hematological disorders featuring iron overload, such as with Thalassemia patients. However, testing these chelators for potential re-purposing on the basis of their antimicrobial activities has yielded poor results as we previously reviewed (Holbein et al. 2021) and now summarize in Table 1. Here, we have compared the antimicrobial activities as reflected by typical Minimum Inhibitory Concentrations (MIC) against members of the three main classes of microbial pathogens, i.e., Gram-positive and Gram-negative bacteria and fungi. Minimum inhibitory concentrations expression on a molar concentration basis (versus weight basis) is important for comparing different chelators given their differing molecular weights. The chemical structures of these chelators are shown in Fig. 2.
Chemical structures of iron chelators. The chemical structures of the three chelators, deferoxamine, deferasirox, and deferiprone as in current clinical use for treating hematological disorders are shown. These three have been shown to have poor antimicrobial activities. Also shown is the structure of the novel chelator DIBI which was specifically developed as an antimicrobial iron withdrawal agent
From Table 1, the problems of hematological chelators as potentially repurposed as antimicrobial therapeutics are fully evident. Deferoxamine is widely known to promote microbial growth which is not surprising given it is itself, a microbial siderophore and because various microbes can utilize it (see xenosiderophores above). It can also be appreciated that high concentrations of deferasirox or deferiprone are required to inhibit microbes. In the case of deferiprone, the required minimum inhibitory concentrations could not be obtained easily in vivo and are in the host-toxic range for the chelator itself. These problems with the currently approved chelators underscore and explain the historical lack of progress in capitalizing on the exploitation of microbial iron dependence using chelators. However, these same problems have informed the design of a new generation of antimicrobial-specific chelating compounds.
We had reviewed (Holbein et al. 2021) the issues of using low molecular weight iron chelators in general, including the clinical hematological types of Table 1, and proposed that the limitations in antimicrobial activities for these are related to their small molecule nature (i.e., molecular weight < 1500 Da), the chemistry of their chelating groups, and their overall iron-binding characteristics. For effective antimicrobial activity of an iron chelator, it is important to ensure that the iron complex formed by the chelators cannot be readily recognized, taken up, or be otherwise accessed by the microbial pathogen.
We have reviewed desirable characteristics for novel anti-infective chelators (Holbein et al. 2021) and here emphasize five key features for useful antimicrobial/anti-infective activity: (1) employing non-siderophore chelating chemistry, i.e., chemically unlike bacterial siderophores in terms of iron chelating component groups; (2) having a molecular size > 1500 Da (i.e., well above the lowest size freely accessible by direct diffusional uptake by both bacterial or host cells; (3) possessing iron binding formation constants as high or higher than either siderophores or current clinical chelators; (4) displaying low toxicity to the infected host, i.e., having high anti-microbial activity but yet a low host toxicity; and (5) of sufficient water solubility to provide flexible formulation and administration options (e.g., intravenous systemic delivery) of the chelator into the infected host to allow it better access and distribution into host extracellular compartments. These various attributes are further discussed below in relation to recently advanced novel chelators.
Hydroxypyridinone, preferred antimicrobial iron chelating ligand
Based on the chemistry of various microbial siderophores (Fig. 1) and the current clinical hematological chelators (Fig. 2), the utility of the hydroxypyridinone chelating group is driven forward. The structure of DIBI a hydroxypyridinone chelator is also shown in Fig. 2 and is discussed below. Hydroxypyridinones combine the chemical characteristics of both hydroxamate and catecholate groups. While catecholate ligands, such as enterobactin and its analogues, have higher stability constants for iron(III) than some hydroxypyridinones, under acidic conditions they are limited by their weak acid nature and require the loss of six protons to bind iron(III). Hydroxypyridinones are monoprotic acids and require the loss of only three protons to form a neutral tris-coordinated iron(III) complex. The structure of the hydroxypyridinone chelate therefore differs significantly from siderophores, and thus, it may not be readily recognized or accessed by bacteria.
Deferiprone (see Table 1, Fig. 2) is a 3-hydroxypyridin-4-one chelator. At pH 7.4, deferiprone is found in its neutral form and the iron(III) complex carries no net charge and can thus penetrate membranes easily to remove potentially toxic iron from tissues (Liu and Hider 2002). The low molecular weight of deferiprone allows its efficient absorption into the intestinal tract and penetration into cells. In addition, it can remove iron(III) from ferritin and transferrin (Dobbin et al. 1993). However, the concentrations needed for antimicrobial activity are very high and would prove toxic for in vivo use (see Table 1). Deferiprone toxicity issues appear related to its ability to enter cells and remove intracellular iron stores, and because being bidentate, it can bind iron in a non-fully coordinated manner under some conditions leaving its bound iron redox reactive for toxic reactive oxygen species generation. Deferiprone’s ability to readily enter cells may also explain its very poor antimicrobial activity as conceivably it could enter microbes providing them intracellular iron.
Hydroxypyridinone polymeric iron chelators
We have previously reviewed various new hydroxypyridinone candidate anti-microbial compounds and the evidence for hexadentate iron binding molecules having greater utility over their corresponding bidentate and tridentate ligands (Holbein et al. 2021). From these attempts, higher molecular weight (i.e., > 1500 Da) has been confirmed as a key aspect for reducing microbial access. In this regard, hydroxypyridinone-containing polymers have emerged as providing desirable antimicrobial benefits. The development of antimicrobial hydroxypyridinone containing polymers addresses the utilities of preferred ligand, increased molecular weight and improved iron capacity. We have also reviewed various approaches to polymer preparation (Holbein et al. 2021) with co-polymerization emerging to be the most versatile approach. Co-polymers offer advantages as to the types and relative amounts of iron binding monomer groups that can be incorporated into the co-polymer while the structural monomer can be selected to provide other desirable characteristics such as water solubility.
To date, two independent approaches have been taken to prepare antimicrobial linear hydroxypyridinone containing co-polymers as illustrated in Fig. 3 The first approach (Zhou et al. 2015a) incorporates monomer ligands with hexadentate binding sites, and their preferred copolymer product 16–4 is shown in Fig. 3. The second approach utilizes bidentate iron binding monomers (Ang et al. 2018; Gumbau-Brisa et al. 2020; Holbein et al. 2011) with a preferred co-polymer product typified by DIBI as shown in Fig. 3.
Structural differences between co-polymer 16–4 (Zhou, Kong, et al. 2015a, b) and co-polymer DIBI (Ang et al. 2018). For co-polymer 16–4, each of the metal binding monomer units used to prepare the co-polymer were capable of hexadentate iron binding. For DIBI, monomer groups capable of only bidentate iron binding were utilized. However, the metal binding characteristics and antimicrobial activities of the resultant co-polymers were strikingly different. The tertiary structure of DIBI appears such that it folds and wraps itself around its bound iron as three neighboring metal binding groups cooperate to complete full hexadentate iron coordination as shown. In contrast, with co-polymer 16–4 cooperation of neighboring monomer groups is not required and the co-polymer likely remains as a linear molecule with iron bound by individual monomer groups as shown. These two different structures result in very different overall iron binding strengths and corresponding antimicrobial activities as reflected in their MIC values for S. aureus. Comparisons of component monomer and resultant copolymer MIC values allows the quantification of the gain in antimicrobial activity achieved with the co-polymer structure over its component metal binding monomer as shown by a measured Polymer Activity Index (PAI). PAI = 1 for 16–4 polymer which indicates no net improvement or degradation of the antimicrobial activity over the free monomer. PAI = 4400 for DIBI, indicating a very strong enhancement of antimicrobial activity of the polymer over the free monomer (figure adapted from Gumbau-Brisa et al. 2020)
Zhou et al. (2015b) co-polymerized a hexadentate 3-hydroxypyridin-4-one monomer with 2-hydroethyl acrylate monomer using azobisisobutyronitrile. The polymer’s iron binding capacity was affected due to the limited incorporation of the monomer during polymerization and the highest achieved was a co-polymer synthesized with an initial monomer mole ratio of 9% and iron capacity of 370 µmol of iron per gram of polymer. The measured log Keq (β3) Fe(III) value of the hexadentate monomer was determined to be 33.6. In addition, the calculated pFe3+ values were similar between the hexadentate monomer and co-polymer, 29.7 and 29.8, respectively; pFe3+ values are related to log Keq (β3) Fe(III) values and represent the negative logarithm of the free ferric iron in solution at pH 7.4 for a defined concentration of ligand (10−5 M) and iron (10–6 M) (Harris et al. 1979). While the log Keq (β3) Fe(III) of the copolymer was not reported by Zhou et al (2015a, b), the similar pFe3+ values indicate that both monomer and polymer species had similar affinity for iron(III) and therefore similar stability constants. This latter aspect is significant in relation to achieving enhanced biological activity over small molecule chelators with iron binding polymers, i.e., where the polymer can be more active as to iron binding and antimicrobial activity than its component monomer (see below).
In the second approach, Ang and co-workers (Ang et al. 2018) described 3-hydroxypyridin-4-one polyvinylpyrrolidone co-polymers. The polymerizations were carried out using commercially available N-vinylpyrrolidone and a bidentate 3-hydroxypyridin-4-one chelating monomer, N-(2-(3-hydroxy-2-methyl-4-oxopyridin-1(4H)-yl)ethyl)methacrylamide (MAHMP), in mild aqueous conditions that gave high yield of the co-polymer. The optimized lead compound, DIBI shown in Fig. 3, has an average content of MAHMP of 24% w/w, with a polymer product recovery of 85% and an iron binding capacity of 338 µmol iron per gram of polymer. The molecular weight of DIBI as measured by gel permeation chromatography was 9 kDa with a polydispersity index of 1.2. DIBI is also highly soluble in water, at up to 70% w/w, without the use of organic solvents (Ang et al. 2018).
Importantly, DIBI showed both a much higher (~ 1000X) iron binding strength and antimicrobial activity in comparison to its MAHMP component monomer. DIBI’s log Keq (β3) Fe(III) = 41.05 for iron is 1000X higher than for its MAHMP monomer with log Keq (β3) Fe(III) = 38.22 (see Table 1, Gumbau-Brisa et al. 2020). Consistent with this stronger iron binding, the minimum inhibitory concentration of DIBI was also approximately 3 orders of magnitude lower in comparison to its component MAHMP monomer (Ang et al. 2018) and in comparison to DFP (Table 1).
DIBI also exhibits a very low toxicity profile (Table 1). No observable toxic effect levels were reached in rats in either 14 day repeated oral or systemic dosing at the highest doses tested (1000 mg/kg/day oral and 200 mg/kg/day intravenous).
DIBI structure teaches a preferred design for antimicrobial polymeric iron chelators
The structural differences between polymer 16–4 and DIBI (see Fig. 3) and their influences on how their iron is bound and therefore their resultant anti-microbial activities suggest these polymers interact with iron very differently in terms of their tertiary structures with their bound iron. The unit hexadentate chelators of polymer 16–4 would likely act independently in binding one iron atom and the resultant metal complexes are strung along the polymer backbone as shown in Fig. 3. From this it can be inferred that the shape of the iron bound structure of 16–4 would not significantly deviate from its iron-free structure. As an aside, in the reported iron-binding dendrimers this is more the case where the chelated iron atoms are rigidly bound to a central scaffold, that render their exposure to the aqueous environment and susceptibility to redox cycling (Zhou et al. 2018, 2014).
In the case of DIBI, the iron binding moieties would aggregate from different locations along the polymer backbone to form the iron tris-chelates, thus forcing the polyvinylpyrrolidone backbone to fold and squeeze against the formed metal complexes. Thus, the polyvinylpyrrolidone backbone would support the internalization of the bound iron, by enveloping it and keeping it physically shielded from the external environment as surrounding the complexed iron structure and thus, possibly hidden from microbes or their siderophores.
The enveloping of the iron complexes due to the “pinching” of the polymer chain and creating a steric barrier around the bound iron has been investigated (Gumbau-Brisa et al. 2020). A globular structure for DIBI was proposed following the binding of iron as opposed to the maintenance of a linear structure with polymer 16–4. Formation of a steric barrier and internalized shielding of the bound iron is supported by the observations that Fe3(DIBI) is resilient to protonation by high concentrations of hydrochloric acid and reduction by the strong reductant, dithionate and which were not observed for solutions of Fe(deferiprone)3 or Fe(MAHMP)3. When DIBI is mixed with Fe(deferiprone)3 with equimolar hydroxypyridinone contents, DIBI was able to steal the iron from deferiprone and shield deferiprone from re-entry to the internal coordination environment of DIBI. As well, DIBI owing to its higher binding affinity was also found to steal iron from transferrin.
The resistance to reductive off-loading of the iron bound to DIBI has biological significance given microbes are capable of reductive iron capture (see section above). DIBI’s ability to off load iron from transferrin (and thus likely lactoferrin as well) is consistent with its in vivo activities of reducing infection, i.e., as to augmenting and bolstering innate host iron withdrawal defenses. In this context DIBI in vivo with transferrin or lactoferrin would provide an augmented capacity of innate ion withdrawal defenses.
Given the above, the structure of DIBI teaches beyond the needs for high molecular weight and water solubility for utility as an antimicrobial polymer. Evidently, physically hiding and withholding iron by a change in morphology upon binding ushers these design aspects forward. The use of polyvinylpyrrolidone as a backbone must be also highlighted, as it is regarded as non-toxic, generally accepted as safe (GRAS), and is ubiquitous in the medical field and it attributes for the high water solubility and low toxicity profile of DIBI.
We have summarized the activity enhancements observed with DIBI’s structure and its measured performance as compared to its component monomer and in comparison, to the 16–4 polymer reported by Zhou (2015). This comparison was possible because both the monomer and the resultant polymer for each were measured for their antimicrobial activities using S. aureus as the test microbe. Table 2 summarizes the polymer activity index as introduced by Gumbau-Brisa et al. (2020) and as determined for both the polymers’ iron binding affinities and their anti-microbial activities. The greatly increased activity (~ 1000X) for DIBI compared to the no net increased activity for polymer 16–4, provide strong supporting chemical characterization evidence for DIBI’s low toxicity and enhanced biological activities as found in animal testing. We have found measurement of polymer activity index has utility for assessing additional polymer leads including from the use of alternate iron-binding monomers and chemical congeners of MAHMP etc. (Ang et al. unpublished results).
Antibiotic resistance and microbial iron needs
Antibiotic resistance occurs from several mechanisms that can arise within a microbe or be acquired from other microbes through the transfer of genetic elements coding for resistance. We and others have previously reviewed these mechanisms (Holbein et al. 2021; Schroeder et al. 2017) with the main mechanisms as summarized in Table 3. It can be appreciated that each of these mechanisms requires microbial growth and/or synthetic activities for their expression and therefore all would have at least some degree of dependence on iron-dependent enzymes. Correspondingly, iron restriction could broadly affect expression of these mechanisms. Examples of key iron-dependent enzymes are ribonucleotide reductase needed for DNA repair/synthesis; cytochrome oxidase needed for ATP energy production and citrate aconitase needed in the tricarboxylic acid (TCA) cycle. Deficiencies in these enzymes could be expected to affect the expression of various antibiotic resistance mechanisms.
We have previously reviewed evidence demonstrating that antibiotic resistance does not lead to a reduced iron requirement of the resistant microbial strain, i.e., relative to its antibiotic sensitive counterpart strain (Holbein et al. 2021). For example, for isogenic S. aureus strains, neither a gain nor a loss of mupirocin resistance resulted in any change in sensitivity to the iron chelator DIBI. In addition, the presence or absence of resistance to broadly different antibiotics (i.e., different chemical classes of antibiotics) for broadly different microbes (both Gram-positive and Gram-negative) did not reflect changes in iron needs based on sensitivity to iron chelators. As well, the requirement for iron as measured for growth was similar for both antibiotic-sensitive and antibiotic-resistant microbes in the cases of C. albicans (Savage et al. 2018), S. aureus (Parquet et al. 2018) and A. baumannii (Parquet et al. 2019). Thus, these various studies establish that the overall iron requirements are similar for both antibiotic-resistant isolates and their antibiotic sensitive counterparts.
Iron chelators suppress infection from antibiotic-resistant microbes
Another key requirement for iron withdrawal agents regarding their potential use as anti-infective alternatives to antibiotics is their ability to suppress infection as caused by antibiotic-resistant microbes, i.e., in addition to demonstrated in vitro activity. This key aspect has seen only limited study to date, but a number of recent studies have demonstrated that novel chelators do possess useful in vivo anti-infective activities against antibiotic-resistant pathogens. Figure 4 summarizes findings from several such studies. The hydroxypyridinone DIBI has been shown to suppress both wound and nares topical infections with antibiotic-resistant S. aureus in mice with two days of topical treatment (i.e., DIBI application to the wound or intranasally, respectively) at approximately 10 µmol/kg per day. The wound infection model utilized for this study was first established to validate fusidic acid efficacy (Kugelberg et al. 2005) and the results with DIBI showed comparable reductions in bacterial burden to that reported by those authors for fusidic acid. The nares carriage model utilized was described by Chhibber et al. (Chhibber et al. 2014) and DIBI was found to reduce nares burdens comparably or better than that reported for mupirocin, noting mupirocin is currently clinically used in man for decolonization of MRSA (Humphreys et al. 2016). Reductions in MRSA bacterial burdens with these discrete infections, i.e., in different body compartments of BALB/C mice, were significant as compared to sham treated controls. Separately, deferasirox with its somewhat improved anti-microbial activity as compared to deferiprone or deferoxamine (see Table 1) has been shown to suppress experimental systemic infection in mice caused by antibiotic-resistant S. aureus MRSA including a vancomycin resistant clinical isolate (Luo et al. 2014). A two-day intravenous treatment course with approximately 10 µmol/kg/day deferasirox resulted in a reduced (although not statistically significant) bacterial burden in both kidneys and spleens of infected mice (Fig. 4). Consistent with these various studies, S. aureus has been shown to upregulate its iron acquisition mechanisms during growth in the nares (Chaves-Moreno et al. 2016) and during systemic infection (Bacconi et al. 2017).
Iron chelators suppress infection from both Gram-positive and Gram-negative antibiotic-resistant bacteria. Mice infected topically with antibiotic-resistant MRSA S. aureus ATCC 43300 on either skin wounds or on their anterior nares were treated with the chelator DIBI as described previously (Parquet et al. 2018) and bacterial burdens after 5 days were compared to sham treated (vehicle treated only) groups. The stars show significant reductions in bacterial burdens compared to the sham treatment. Mice infected intranasally with A. baumannii antibiotic-resistant isolate LAC-4 to initiate pneumonia followed by septicemia as reported by Parquet et al. (2019) were treated intranasally once with DIBI or sham treated with only vehicle and bacterial burdens after 24 h infection were determined for both lungs and spleens. The stars show significant reductions in bacterial burdens compared to the sham treatment. In other testing reported by Luo et al (2014) mice were injected intravenously with S. aureus MRSA antibiotic-resistant isolate LAC300 to initiate a systemic infection and bacterial burdens in kidneys and spleens were compared in groups treated with either deferasirox (DEF) or placebo (sham treated). In those studies, DEF tended to reduce bacterial burdens but the results were not found to be statistically significant (the results as graphed are based only on estimates of bacterial burdens taken from the Luo publication). All these various infection studies show that iron chelators on their own can suppress infection for various antibiotic-resistant bacterial infections
In a separate study of experimental pneumonia/septicemia in BALB/C mice caused by intranasal introduction of a hyper virulent antibiotic-resistant isolate of A. baumannii, DIBI reductions in A. baumannii bacterial burdens in both the lung and spleen (see Fig. 4) were comparable to those obtained with tigecycline an antibiotic to which this isolate is highly susceptible (Parquet et al. 2019). Importantly, A. baumannii has also been shown to upregulate its iron acquisition systems during infection (Gaddy et al. 2012) indicating iron supply is important to this infection.
These various infection studies with antibiotic-resistant bacteria demonstrate that growth of these bacteria during infection is at least partially iron restricted by host iron withdrawal defenses causing these pathogens to upregulate their iron acquisition mechanisms and in turn, ensuring their sensitivity to iron sequestering chelator addition. Given the findings that iron chelators such as deferasirox or DIBI suppress infection, it seems reasonable to conclude that these chelators play a role in bolstering natural innate host iron withdrawal defense mechanisms to various other infections.
Iron chelators enhance antibiotic activity
A very important aspect in considering the use of iron withdrawal chelators as alternatives to antibiotics is that these, if co-administered with antibiotics, should not interfere with antibiotic efficacy. On the other hand, a clear benefit would be obtained if chelators through withdrawing microbial iron supplies improved the activities of antibiotics including against antibiotic-resistant isolates. These desired attributes have now been demonstrated in various in vitro and in vivo studies.
Some of the first evidence that iron withdrawal agents could positively affect antibiotic action for antibiotic-resistant isolates was in relation to lactoferrin which is a host defense iron binding transferrin-class protein found in exocrine secretions such as tears and in colostrum. Diarra et al. (Diarra et al. 2002) demonstrated that lactoferrin synergistically improved the activity of penicillin including against penicillin resistant isolates of S. aureus isolates recovered from bovine mastitis and this use for lactoferrin had shown clinical promise (Lacasse et al. 2008). More recently, lactoferrin has also been shown to enhance amphotericin B activity against yeast infection, where the combination therapy significantly prolonged the survival of Galleria mellonella wax moth larvae infected with Candida albicans or Cryptococcus neoformans decreasing fungal burden up to 25-fold (Fernandes et al. 2020).
However, clinical use of lactoferrin (or other transferrin class proteins) has yet to prove practical, possibly due to issues related to their larger scale production, collection and purification of these animal proteins for use in man (or other animals) as well as issues related to their administration. In contrast, synthetic chelators offer strong potential for large scale cost-effective production, robust quality assurance and for those that are soluble in aqueous media (such as DIBI, see below), flexible formulation and administration options, including systemic delivery.
To conclude there is synergy of activity between an iron chelator (or any other adjunct) with an antibiotic one standard test is the time/kill kinetic assay as typically applied to test bactericidal antibiotics. In these tests, a somewhat reduced concentration of antibiotic or the test chelator are typically used separately, i.e., so that each agent on its own does not provide complete killing when added to a relatively high number of rapidly growing (log-phase) bacterial cells. This testing approach then allows a better assessment of the combined agent affects.
It is important to note that while many antibiotics are bactericidal, chelators are fundamentally different as to mode of action and are not directly bactericidal in such assays. Rather, chelators act to deny access to iron as essential to microbe growth, and therefore, microbial killing by these agents is delayed as compared to bactericidal antibiotics. Bacterial death from a chelator would ensue due to eventual iron insufficiency leading to apoptosis. An additional method to assess agent synergy is to perform checkerboard cross-titrations of the antibiotic and chelator in MIC test plates so as to measure the Fractional Inhibition Concentration Index or FICI; if the FICI is ≤ 0.5, synergy between the antibiotic and chelator is evident, and if > 1.0, the agent pairing is antagonistic. Deferasirox and DIBI (see Table 1 for comparative characteristics) and Apo-Tf (i.e., serum transferrin stripped of its iron) have been shown to not interfere but to synergistically enhance antibiotic activity with various antibiotic-resistant bacteria as discussed below.
To illustrate the results and significance of kinetic kill synergy studies, an example using gentamicin with A. baumannii is shown in Fig. 5. Note again, for this testing, gentamicin was added at 1X MIC and on its own provided only partial initial killing followed by strong recovery growth to near control levels of bacterial colony forming units (CFU)/mL by 24 h.

adapted from Parquet et al. 2019)
Example of a time kill kinetic assay showing chelator enhancement of antibiotic killing and prevention of survivor re-growth. A. baumannii ATCC 17978 which is sensitive to the aminoglycoside Gentamicin (GEN) (Minimum Inhibitory Concentration (MIC) = 1 µg/mL) was exposed to 0.5 µg/mL GEN (i.e., 1/2 MIC) and this caused only partial killing of the population followed by rapid recovery growth that reached near untreated control Colony Forming Units (CFU) levels by 24 h of exposure. The DIBI concentration used in this experiment was 20 µg/mL and this on its own did not affect total CFU by 24 h. It should be noted that a large rapidly growing iron replete bacterial population had been introduced at 0 h and gross effects of DIBI on bacterial numbers would not be expected under these test conditions. However, this DIBI treatment exhibited effects of iron withdrawal as evident when DIBI was applied in combination with GEN. The GEN/DIBI combination resulted in extensive and continued bacterial killing with no detectable CFU remaining at 24 h exposure (results
The chelator DIBI was also tested at a low concentration, i.e., that was insufficient on its own to fully arrest growth of the rapidly dividing log phase cells. However, the combination of gentamicin + DIBI resulted in a > 8 log reduction in bacterial CFU by 24 h. The 24-h measurement point in a time/kill assay has significance as it is generally accepted that synergy of two anti-infectives is confirmed if there is ≥ 2 log CFU reduction provided by the combination by 24 h incubation, i.e., as compared to untreated control or to individual antibiotic or adjunct agent related CFU reductions/increases at 24 h.
Similar patterns of time/kill responses or FICI findings demonstrating synergy have been reported for other bacterial species and for other antibiotics in combination with various iron withdrawal agents including deferasirox, DIBI or Apo-transferrin or Apo-lactoferrin (bovine milk lactoferrin stripped of its iron) as summarized in Table 4. In all these cases, deferasirox, DIBI, Apo-transferrin, or Apo-lactoferrin did not impede but typically improved antibiotic activity.
The phenomenon of microbial recovery growth following an antibiotic exposure as shown for gentamicin alone in Fig. 5 is typically observed with various antibiotics and for various microbes, and this phenomenon has important clinical relevance in relation to the development of antibiotic resistance. Antibiotics are typically administered at a relatively high dose, i.e., multiples of MIC, but later during a treatment course, due to antibiotic clearance etc., concentrations can wane. Thus, if bacterial survivors with some degree of antibiotic resistance are present under these conditions, they survivors can more likely regrow as a population with resistance to the antibiotic that had been applied. The condition of suboptimal antibiotic concentration strongly favors selection and outgrowth of antibiotic-resistant survivors.
On the basis of various results, we have summarized herein new therapeutics that function to withdraw microbial iron have high potential to work in conjunction with conventional antibiotics to improve their activity, including against antibiotic-resistant bacteria. The findings that the host defense protein, transferrin, increased ciprofloxacin killing and also prevented regrowth of ciprofloxacin-resistant survivors (Ambrose et al. 2019) provide additional support for using synthetic chelators. Such chelators can act to assist or bolster innate iron withdrawal defenses as provided by serum transferrin or closely related lactoferrin. Consistent with this, time-kill recovery growth assays in our laboratory have shown that ciprofloxacin at 2 X MIC provided a substantial initial killing of S. aureus but with continued incubation a progressively enriched population of ciprofloxacin-resistant survivor clones appeared, reaching near 100% ciprofloxacin resistant clones by 96 h (Allan and Holbein, unpublished results). When randomly selected surviving clones exposed to ciprofloxacin alone were recovered and re-tested for MIC, they had an MIC increase from 0.125 to 1 µg/mL (eightfold), whereas surviving clones exposed to ciprofloxacin and DIBI showed no change in MIC to either agent. This suggests that DIBI may be able to prevent a spontaneous mutation or its expression that leads to reduced susceptibility to ciprofloxacin; this interesting finding requires further study.
In addition, the prospect for resistance development to iron withdrawal agents is very low given iron is an essential nutrient. Testing with DIBI at sub-inhibitory concentrations under cultural conditions that readily promote antibiotic resistance development (i.e., when antibiotic present) has failed to show any resistance development to DIBI even after repeated culture passages under these conditions (Allan et al. 2020).
Conclusion
The essential and irreplaceable iron requirements of antibiotic-resistant microbial pathogens are similar to their antibiotic sensitive counterparts. As well, iron availability to microbes is restricted by the host during infection. These features provide foundations for using iron withdrawal by chelators as a therapeutic means to both suppress pathogen growth in vivo and address antibiotic-resistant infection. New generation synthetic purpose-designed antimicrobial chelators especially those with 3-hydroxypyridin-4-one functionality have excellent potential to become both stand-alone anti-infectives and adjunct agents for use with conventional antibiotics. Among these, the hydroxypyridinone containing copolymer, DIBI, has seen substantial development, successful proof-of-concept testing, it has a low host toxicity and it complements the host’s natural iron withdrawal defense mechanisms to suppress infection. In addition, iron withdrawal by both host iron-withdrawing defense proteins and chelators including DIBI, improves antibiotic microbial killing activity and very importantly, suppresses antibiotic resistance development in antibiotic-exposed microbes.
These various attributes open the potential for using iron withdrawal agents as new host-directed therapeutic agents. Host-directed therapies do not rely on narrowly-targeted mechanisms that have been the major problem to date with employing antibiotics. Iron withdrawal antimicrobial therapeutics have potential for use as broad spectrum medications for infection as we have previously outlined in more detail (Holbein et al. 2021).
DIBI, owing to its unique structure and mechanism for binding and sequestering iron, has demonstrated enhanced capabilities which support its potential for addressing at least two major unmet needs as outlined below.
Early intervention treatment in serious infection/sepsis
Clinical treatment of sepsis from serious infection is challenging for three main reasons. First, antibiotic resistance of bacteria is increasing and along with this, the incidence of sepsis is mounting as well. More concerning, there is no currently approved therapy available to treat the dysregulated immune response of sepsis. Thirdly, treatment for sepsis is often limited to supportive treatments with these initiated later during the infection course after the dysregulated immune response has already progressed, possibly to its irreversibility. With the only ever approved drug for sepsis therapy (drotrecogin alpha (activated)—Xigris®) having been withdrawn from the market in 2011 (Martí-Carvajal et al. 2011), there is a large unmet need to improve the outcome of this deadly disease.
A host-directed therapeutic approach using new antimicrobial iron chelators such as DIBI has potential to address this need. Preclinical data suggest promise for including iron sequestration agents early in the therapy of severe infection and sepsis to boost host innate iron withdrawal defenses, reduce bacterial growth, overcome antibiotic resistance and also dampen hyper-inflammatory Reactive Oxygen Species (ROS) production, reducing tissue damage. Because DIBI has broad spectrum antimicrobial activity and does not interfere with but actually improves antibiotic efficacy it could be applied early in suspected serious infections, possibly at or even ahead of antibiotic administration, i.e., should this be delayed pending microbiological culture confirmation and antibiotic susceptibility screening.
The concept of reducing iron levels by iron sequestration agents often raises concerns by physicians and their patients, since physiologic iron levels are generally considered to be required and optimal for health. However, we believe this may not apply for life-threatening conditions such as sepsis especially given dysregulated iron metabolism has now been implicated in sepsis. Based on the recent preclinical data, the combination of antibiotics and iron chelators has synergistic effects to fight infection and improve the immune response in sepsis (Fokam et al. 2020b; Lehmann et al. 2021). In addition, there is no doubt that under iron overload conditions, iron chelation is beneficial to reduce the risk of infections in humans (Schaible and Kaufmann 2004). Furthermore, because of the increased infection risk in the surgical patient or those compromised such as on ventilator support, we encourage further investigation leading to clinical safety/efficacy trials of iron sequestration agents in the prophylaxis regimen for such infections of compromised patients.
Adjuncts with antibiotics to treat antimicrobial resistance infection
It is now clear based on a variety of studies that iron chelators do not interfere with antibiotic activity but in fact improve activity of antibiotics from various chemical classes of antibiotics. Possibly most important is that iron chelators when with an antibiotic, suppress outgrowth of antibiotic-resistant survivors and therefore, act to suppress resistance development. New adjunct formulations of existing antibiotics with antimicrobial iron withdrawal agents such as DIBI could serve to extend the usage life of conventional antibiotics and address treatment needs for antibiotic-resistant infections. In this regard, it is noteworthy that testing of DIBI has shown no evidence for resistance development to DIBI.
Abbreviations
- DFP:
-
Deferiprone
- DEF:
-
Deferasirox
- DFO:
-
Deferoxamine
- DIBI:
-
Depriving iron from bacterial infections
- ROS:
-
Reactive oxygen species
- ABC:
-
ATP-binding cassette
- ATP:
-
Adenosine triphosphate
- Fe-Tf:
-
Transferrin iron
- Lf:
-
Lactoferrin
- LPS:
-
Lipopolysaccharide
- LTA:
-
Lipoteichoic acid
- IL-6:
-
Interlukin-6
- TLR4:
-
Toll-like receptor 4
- NF-kB:
-
Nuclear factor-kB
- TNF-α:
-
Tumor necrosis factor-alpha
- FDA:
-
Food and Drug Administration
- PBP:
-
Penicillin-binding proteins
- MIC:
-
Minimum inhibitory concentration
- MAHMP:
-
N-(2-(3-Hydroxy-2-methyl-4-oxopyridin-1(4H)-yl)ethyl)methacrylamide
- GRAS:
-
Generally accepted as safe
- DNA:
-
Deoxyribonucleic acid
- TCA cycle:
-
Tricarboxylic acid cycle
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- CFU:
-
Colony forming units
- FICI:
-
Fractional inhibition concentration index
References
Afeltra A, Caccavo D, Ferri GM, Addessi MA, De Rosa FG, Amoroso A, Bonomo L (1997) Expression of lactoferrin on human granulocytes: analysis with polyclonal and monoclonal antibodies. Clin Exp Immunol 109(2):279–285. https://doi.org/10.1046/j.1365-2249.1997.4351333.x
Allan DS, Parquet MC, Savage KA, Holbein BE (2020) Iron Sequestrant DIBI, a potential alternative for nares decolonization of methicillin-resistant Staphylococcus aureus, is anti-infective and inhibitory for mupirocin-resistant isolates. Antimicrob Agents Chemother 64(3):e02353-e12319. https://doi.org/10.1128/AAC.02353-19
Altamura S, Kopf S, Schmidt J, Müdder K, da Silva AR, Nawroth P, Muckenthaler MU (2017) Uncoupled iron homeostasis in type 2 diabetes mellitus. J Mol Med 95(12):1387–1398. https://doi.org/10.1007/s00109-017-1596-3
Amaral L, Martins A, Spengler G, Molnar J (2014) Efflux pumps of Gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front Pharmacol 4:168. https://doi.org/10.3389/fphar.2013.00168
Ambrose PG, VanScoy BD, Luna BM, Yan J, Ulhaq A, Nielsen TB, Rudin S, Hujer K, Bonomo RA, Actis L, Skaar E, Spellberg B (2019) Apotransferrin in combination with ciprofloxacin slows bacterial replication, prevents resistance amplification, and increases antimicrobial regimen effect. Antimicrob Agents Chemother 63(5):e00112-00119. https://doi.org/10.1128/AAC.00112-19
Ang MTC, Gumbau-Brisa R, Allan DS, McDonald R, Ferguson MJ, Holbein BE, Bierenstiel M (2018) DIBI, a 3-hydroxypyridin-4-one chelator iron-binding polymer with enhanced antimicrobial activity. MedChemComm 9:1206–1212. https://doi.org/10.1039/c8md00192h
Bacconi M, Haag AF, Chiarot E, Donato P, Bagnoli F, Delany I, Bensi G (2017) In vivo analysis of Staphylococcus aureus-infected mice reveals differential temporal and spatial expression patterns of fhuD2. Infect Immun 85(10):e00270-e1217. https://doi.org/10.1128/IAI.00270-17
Badeli H, Baghersalimi A, Eslami S, Saadat F, Rad AH, Basavand R, Papkiadeh SR, Darbandi B, Kooti W, Peluso I (2019) Early kidney damage markers after deferasirox treatment in patients with thalassemia major: a case-control study. Oxid Med Cell Longev 2019:5461617. https://doi.org/10.1155/2019/5461617
Bairwa G, Hee Jung W, Kronstad JW (2017) Iron acquisition in fungal pathogens of humans. Metallomics 9(3):215–227. https://doi.org/10.1039/c6mt00301j
Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, Martinez-Urtaza J (2018) Vibrio spp. infections. Nat Rev Dis Primers 4(1):1–19. https://doi.org/10.1038/s41572-018-0005-8
Banin E, Lozinski A, Brady KM, Berenshtein E, Butterfield PW, Moshe M, Chevion M, Greenberg EP, Banin E (2008) The potential of desferrioxamine-gallium as an anti-Pseudomonas therapeutic agent. Proc Natl Acad Sci USA 105(43):16761–16766. https://doi.org/10.1073/pnas.0808608105
Barrasa-Villar JI, Aibar-Remón C, Prieto-Andrés P, Mareca-Doñate R, Javier M-L (2017) Impact on morbidity, mortality, and length of stay of hospital-acquired infections by resistant microorganisms. Clin Infect Dis 65(4):644–652. https://doi.org/10.1093/cid/cix411
Barton JC, Acton RT (2019) Hepcidin, iron, and bacterial infection. Vitam Horm 110:223–242. https://doi.org/10.1016/bs.vh.2019.01.011
Bengtsson-Palme J, Kristiansson EDG, Larsson J (2018) Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev 42:68–80. https://doi.org/10.1093/femsre/fux053
Bilitewski U, Blodgett JAV, Duhme-Klair AK, Dallavalle S, Laschat S, Routledge A, Schobert R (2017) Chemical and biological aspects of nutritional immunity-perspectives for new anti-infectives that target iron uptake systems. Angew Chem Int Ed Engl 56(46):14360–14382. https://doi.org/10.1002/anie.201701586
Bonomo RA (2019) Cefiderocol: a novel siderophore cephalosporin defeating carbapenem-resistant pathogens. Clin Infect Dis 69(Suppl 7):S519–S520. https://doi.org/10.1093/cid/ciz823
Brandtner A, Tymoszuk P, Nairz M, Lehner GF, Fritsche G, Vales A, Falkner A, Schennach H, Theurl I, Joannidis M, Weiss G, Pfeifhofer-Obermair C (2020) Linkage of alterations in systemic iron homeostasis to patients’ outcome in sepsis: a prospective study. J Intensive Care 8:76. https://doi.org/10.1186/s40560-020-00495-8
Britigan BE, Hayek MB, Doebbeling BN, Fick RBJ (1993) Transferrin and lactoferrin undergo proteolytic cleavage in the Pseudomonas aeruginosa-infected lungs of patients with cystic fibrosis. Infect Immun 61(12):5049–5055. https://doi.org/10.1128/iai.61.12.5049-5055.1993
Budzikiewicz H (2001) Siderophore-antibiotic conjugates used as trojan horses against Pseudomonas aeruginosa. Curr Top Med Chem 1(1):73–82. https://doi.org/10.2174/1568026013395524
Caza M, Kronstad JW (2013) Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol 3:80. https://doi.org/10.3389/fcimb.2013.00080
Chaves-Moreno D, Wos-Oxley ML, Jáuregui R, Medina E, Oxley APA, Pieper DH (2016) Exploring the transcriptome of Staphylococcus aureus in its natural niche. Nat Sci Rep 6:33174. https://doi.org/10.1038/srep33174
Chhibber S, Gupta P, Kaur S (2014) Bacteriophage as effective decolonising agent for elimination of MRSA from anterior nares of BALB/c mice. BMC Microbiol 14:212
Chitambar CR (2010) Medical applications and toxicities of gallium compounds. Int J Environ Res Public Health 7(5):2337–2367. https://doi.org/10.3390/ijerph7052337
Chitambar CR (2016) Gallium and its competing roles with iron in biological systems. Biochim Biophys Acta 1863(8):2044–2053. https://doi.org/10.1016/j.bbamcr.2016.04.027
Chitambar CR (2017) The therapeutic potential of iron-targeting gallium compounds in human disease: from basic research to clinical application. Pharmacol Res 115:56–64. https://doi.org/10.1016/j.phrs.2016.11.009
Cronin SJF, Woolf CJ, Weiss G, Penninger JM (2019) The role of iron regulation in immunometabolism and immune-related disease. Front Mol Biosci 6:116. https://doi.org/10.3389/fmolb.2019.00116
Czaplewski L, Bax R, Clokie M, Dawson M, Fairhead H, Fischetti VA, Foster S, Gilmore BF, Hancock REW, Harper D, Henderson IR, Hilpert K, Jones BV, Kadioglu A, Knowles D, Ólafsdóttir S, Payne D, Projan S, Shaunak S, Silverman J, Thomas CM, Trust TJ, Warn P, Rex JH (2016) Alternatives to antibiotics—a pipeline portfolio review. Lancet Infect Dis 16:239–251. https://doi.org/10.1016/S1473-3099(15)00466-1
de Leseleuc L, Harris G, KuoLee R, Xu HH, Chen W (2014) Serum resistance, gallium nitrate tolerance and extrapulmonary dissemination are linked to heme consumption in a bacteremic strain of Acinetobacter baumannii. Int J Med Microbiol 304(3–4):360–369. https://doi.org/10.1016/j.ijmm.2013.12.002
Diarra MS, Petitclerc D, Lacasse P (2002) Effect of lactoferrin in combination with penicillin on the morphology and the physiology of Staphylococcus aureus isolated from bovine mastitis. J Dairy Sci 85(5):1141–1149. https://doi.org/10.3168/jds.s0022-0302(02)74176-3
Dickson KB, Zhou J (2020) Role of reactive oxygen species and iron in host defense against infection. Front Biosci Landmark 25:1600–1616. https://doi.org/10.2741/4869
Dobbin PS, Hider RC, Hall AD, Taylor PD, Sarpong P, Porter JB (1993) Synthesis, physicochemical properties, and biological evaluation of N-substituted 2-alkyl-3-hydroxy-4(1″)-pyridinones: orally active iron chelators with clinical potential. J Med Chem 36(17):2448–2458. https://doi.org/10.1021/jm00069a002
Ellermann M, Arthur JC (2017) Siderophore-mediated iron acquisition and modulation of host-bacterial interactions. Free Radic Biol Med 105:68–78. https://doi.org/10.1016/j.freeradbiomed.2016.10.489
Fernandes KE, Weeks K, Carter DA (2020) Lactoferrin is broadly active against yeasts and highly synergistic with amphotericin B. Antimicrob Agents Chemother 64(5):e02284-e12219. https://doi.org/10.1128/AAC.02284-19
Fokam D, Hoskin D (2020) Instrumental role for reactive oxygen species in the inflammatory response. Front Biosci Landmark 25(6):1110–1119. https://doi.org/10.2741/4848
Fokam D, Aali M, Dickson K, Scott C, Holbein BE, Zhou J, Lehmann C (2020a) The novel iron chelator, DIBI, attenuates inflammation and improves outcome in colon ascendens stent peritonitis-induced experimental sepsis. Clin Hemorheol Microcirc 76(2):241–261. https://doi.org/10.3233/ch-209207
Fokam D, Dickson K, Kamali K, Holbein BE, Colp P, Stueck A, Zhou J, Lehmann C (2020b) Iron chelation in murine models of systemic inflammation induced by gram-positive and gram-negative toxins. Antibiotics (Basel, Switzerland) 9(6):283. https://doi.org/10.3390/antibiotics9060283
Foley TL, Simeonov A (2012) Targeting iron assimilation to develop new antibacterials. Expert Opin Drug Discov 7(9):831–847. https://doi.org/10.1517/17460441.2012.708335
Friedman ND, Temkin E, Carmeli Y (2016) The negative impact of antibiotic resistance. Clin Microbiol Infect 22:416–422. https://doi.org/10.1016/j.cmi.2015.12.002
Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA (2012) Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars and mice. Infect Immun 80:1015–1024. https://doi.org/10.1128/IAI.06279-11
Ganz T (2018) Iron and infection. Int J Hematol 107:7–15. https://doi.org/10.1007/s12185-017-2366-2
Ganz T, Nemeth E (2015) Iron homeostasis in host defence and inflammation. Nat Rev Immunol 15:500–510. https://doi.org/10.1038/nri3863
Gumbau-Brisa R, Ang MTC, Holbein BE, Bierenstiel M (2020) Enhanced Fe3+ binding through cooperativity of 3-hydroxypyridin-4-one groups within a linear co-polymer: wrapping effect leading to superior antimicrobial activity. Biometals 33(6):339–351. https://doi.org/10.1007/s10534-020-00253-1
Gupta P, Sarkar S, Das B, Bhattacharjee S, Tribed P (2016) Biofilm, pathogenesis and prevention—a journey to break the wall: a review. Arch Microbiol 198:1–15. https://doi.org/10.1007/s00203-015-1148-6
Harris WR, Carrano CJ, Cooper SR, Sofen SR, Avdeef AE, McArdle JV, Raymond KN (1979) Coordination chemistry of microbial iron transport compounds. 19. Stability constants and electrochemical behavior of ferric enterobactin and model complexes. J Am Chem Soc 101(20):6097–6104. https://doi.org/10.1021/ja00514a037
Holbein B (1980) Iron-controlled infection with Neisseria meningitidis in mice. Infect Immun 29(3):886–891
Holbein BE (1981) Enhancement of Neisseria meningitidis infection in mice by addition of iron bound to transferrin. Infect Immun 34(1):120–125
Holbein BE, Feng M, Huber AL, Kidby DK (2011) Polymeric metal chelating compositions and methods of preparing same for controlling growth and activities of living cells and organisms. International patent no. WO2012167368 A1
Holbein BE, Ang MTC, Allan DS, Chen W, Lehmann C (2021) Exploiting the Achilles’ heel of iron dependence in antibiotic resistant bacteria with new antimicrobial iron withdrawal agents. In: Panwar H, Sharma C, Lichtfouse E (eds) Sustainable agriculture reviews, vol 49. Springer, Cham. https://doi.org/10.1007/978-3-030-58259-3_8
Holden VI, Bachman MA (2015) Diverging roles of bacterial siderophores during infection. Metallomics 7(6):986–995. https://doi.org/10.1039/c4mt00333k
Humphreys H, Becker K, Dohmen PM, Petrosillo N, Spencer M, van Rijen M, Wechsler-Fordos A, Pujol M, Dubouix A, Garau J (2016) Staphylococcus aureus and surgical site infections: benefits of screening and decolonization before surgery. J Hosp Infect 94:295–304. https://doi.org/10.1016/j.jhin.2016.06.011
Ibrahim AS, Gebremariam T, French SW, Edwards JE Jr, Spellberg B (2010) The iron chelator deferasirox enhances liposomal amphotericin B efficacy in treating murine invasive pulmonary aspergillosis. J Antimicrob Chemother 65(2):289–292. https://doi.org/10.1093/jac/dkp426
Islam S, Jarosch S, Zhou J, Parquet MC, Toguri JT, Colp P, Holbein BE, Lehmann C (2016) Anti-inflammatory and anti-bacterial effects of iron chelation in experimental sepsis. J Surg Res 200(1):266–273. https://doi.org/10.1016/j.jss.2015.07.001
Jandu AS, Vidgeon S, Ahmed N (2018) Anaemia and transfusion triggers in critically ill patients—what we have learnt thus far. J Intensive Care Soc 20(4):284–289. https://doi.org/10.1177/1751143718783615
Johnson EE, Wessling-Resnick M (2012) Iron metabolism and the innate immune response. Microbes Infect 14(3):207–216. https://doi.org/10.1016/j.micinf.2011.10.001
Johnstone TC, Nolan EM (2015) Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans 44(14):6320–6339. https://doi.org/10.1039/c4dt03559c
Katz L, Baltz RH (2016) Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol 43:155–176. https://doi.org/10.1007/s10295-015-1723-5
Kelson AB, Carnevali M, Truong-Le V (2013) Gallium-based anti-infectives: targeting microbial iron-uptake mechanisms. Curr Opin Pharmacol 13(5):707–716. https://doi.org/10.1016/j.coph.2013.07.001
Khan A, Singh P, Srivastava A (2018) Synthesis, nature and utility of universal iron chelator—siderophore: a review. Microbiol Res 212–213:103–111. https://doi.org/10.1016/j.micres.2017.10.012
Kugelberg E, Norstro T, Petersen TK, Duvold T, Andersson DI, Hughes D (2005) Establishment of a superficial skin infection model in mice by using Staphylococcus aureus and Streptococcus pyogenes. Antimicrob Agents Chemother 49:3435–3441. https://doi.org/10.1128/AAC.49.8.3435-3441.2005
Lacasse P, Lauzon K, Diarra MS, Petitclerc D (2008) Utilization of lactoferrin to fight antibiotic-resistant mammary gland pathogens. J Anim Sci 86:66–71. https://doi.org/10.2527/jas.2007-0216
Larsen R, Gozzelino R, Jeney V, Tokaji L, Bozza FA, Japiassú AM, Bonaparte D, Cavalcante MM, Chora Â, Ferreira A, Marguti I, Cardoso S, Sepúlveda N, Smith A, Soares MP (2010) A central role for free heme in the pathogenesis of severe sepsis. Sci Transl Med 2(51):51–71. https://doi.org/10.1126/scitranslmed.3001118
Lehmann C, Aali M, Zhou J, Holbein BE (2021) Comparison of treatment effects of different iron chelators in experimental models of sepsis. Life (Basel, Switzerland) 11(1):57. https://doi.org/10.3390/life11010057
Letendre ED, Holbein BE (1983) Turnover in the transferrin iron pool during the hypoferremic phase of experimental neisseria meningitidis infection in mice. Infect Immun 39(1):50–59
Letendre ED, Holbein BE (1984) Mechanism of impaired iron release by the reticuloendothelial system during the hypoferremic phase of experimental neisseria meningitidis infection in mice. Infect Immun 44(2):320–325
Liu ZD, Hider RC (2002) Design of clinically useful iron(III)-selective chelators. Med Res Rev 22(1):26–64. https://doi.org/10.1002/med.1027
Liu X, Deng S, Huang J, Huang Y, Zhang Y, Yan Q, Wang Y, Li Y, Sun C, Xu J (2017) Dissemination of macrolides, fusidic acid and mupirocin resistance among Staphylococcus aureus clinical isolates. Oncotarget 8:58086–58097. https://doi.org/10.18632/oncotarget.19491
Lubelski J, Konings WN, Driessen AJM (2007) Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev 71:463–476. https://doi.org/10.1128/MMBR.00001-07
Luepke KH, Suda KJ, Boucher H, Russo RL, Bonney MW, Hunt TD, Mohr JF (2017) Past, Present, and future of antibacterial economics: increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy 37:71–84. https://doi.org/10.1002/phar.1868
Luo G, Spellberg B, Gebremariam T, Lee H, Xiong YQ, French SW, Bayer A, Ibrahim AS (2014) Combination therapy with iron chelation and vancomycin in treating murine staphylococcemia. Eur J Clin Microbiol Infect Dis 33:845–851. https://doi.org/10.1007/s10096-013-2023-5
Manyi-Loh C, Mamphweli S, Meyer E, Okoh A (2018) Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23(4):795. https://doi.org/10.3390/molecules23040795
Martí-Carvajal AJ, Solà I, Lathyris D, Cardona AF (2011) Human recombinant activated protein C for severe sepsis. Cochrane Database Syst Rev 4:CD004388
Mollmann U, Heinisch L, Bauernfeind A, Kohler T, Ankel-Fuchs D (2009) Siderophores as drug delivery agents: application of the “Trojan Horse” strategy. Biometals 22(4):615–624. https://doi.org/10.1007/s10534-009-9219-2
Movassaghi M, van der Donk WA (2018) Synthesis of antibiotics and related molecules. J Org Chem 83:6826–6828. https://doi.org/10.1021/acs.joc.8b01330
Nairz M, Dicht S, Schroll A, Haschka D, Tymoszuk P, Theurl I, Weiss G (2018) Iron and innate antimicrobial immunity—depriving the pathogen, defending the host. J Trace Elem Med Biol 48:118–133. https://doi.org/10.1016/j.jtemb.2018.03.007
Naseer S, Weinstein EA, Rubin DB, Suvarna K, Wei X, Higgins K, Goodwin A, Jang SH, Iarikov D, Farley J, Nambiar S (2020) US Food and Drug Administration (FDA): benefit-risk considerations for cefiderocol (Fetroja(R)). Clin Infect Dis. https://doi.org/10.1093/cid/ciaa1799
Neupane GP, Kim DM (2010) In vitro time-kill activities of ciprofloxacin alone and in combination with the iron chelator deferasirox against Vibrio vulnificus. Eur J Clin Microbiol Infect Dis 29(4):407–410. https://doi.org/10.1007/s10096-010-0875-5
Palmer LD, Skaar EP (2016) Transition metals and virulence in bacteria. Annu Rev Genet 50:67–91. https://doi.org/10.1146/annurev-genet-120215-035146
Paoli CJ, Reynolds MA, Sinha M, Gitlin M, Crouser E (2018) Epidemiology and costs of sepsis in the united states—an analysis based on timing of diagnosis and severity level. Crit Care Med 46(12):1889–1897. https://doi.org/10.1097/CCM.0000000000003342
Parquet MC, Savage KA, Allan DS, Davidson RJ, Holbein BE (2018) Novel iron-chelator DIBI inhibits Staphylococcus aureus growth, suppresses experimental MRSA infection in mice and enhances the activities of diverse antibiotics in vitro. Front Microbiol 9:1811. https://doi.org/10.3389/fmicb.2018.01811
Parquet MC, Savage KA, Allan DS, Ang MTC, Chen W, Logan SM, Holbein BE (2019) Antibiotic resistant Acinetobacter baumannii is susceptible to the novel iron-sequestering anti-infective DIBI in vitro and in experimental pneumonia in mice. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.00855-19
Pearl RG (2014) Erythropoietin and organ protection: lessons from negative clinical trials. Crit Care 18(5):526. https://doi.org/10.1186/s13054-014-0526-9
Post SJ, Shapiro JA, Wuest WM (2019) Connecting iron acquisition and biofilm formation in the ESKAPE pathogens as a strategy for combatting antibiotic resistance. MedChemComm 10(4):505–512. https://doi.org/10.1039/C9MD00032A
Powers JH III (2020) Scientific evidence, regulatory decision making, and incentives for therapeutics in infectious diseases: the example of cefiderocol. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa1795
Prasad T, Chandra A, Mukhopadhyay CK, Prasad R (2006) Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob Agents Chemother 50(11):3597–3606. https://doi.org/10.1128/AAC.00653-06
Proctor R, Kahl B, von Eiff C, Vaudaux P, Daniel P, Lew D, Peters G (1998) Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin Infect Dis 27:S68-74. https://doi.org/10.1086/514906
Proctor R, Kriegeskorte A, Kahl B, Becker K, Löffler B, Peters G (2014) Staphylococcus aureus small colony variants (SCVs): a roadmap for the metabolic pathways involved in persistent infections. Front Cell Infect Microbiol 4:1–8. https://doi.org/10.3389/fcimb.2014.00099
Rangel-Vega A, Bernstein LR, Mandujano-Tinoco EA, Garcia-Contreras SJ, Garcia-Contreras R (2015) Drug repurposing as an alternative for the treatment of recalcitrant bacterial infections. Front Microbiol 6:282. https://doi.org/10.3389/fmicb.2015.00282
Richter K, Thomas N, Zhang G, Prestidge C, Coenye T, Wormald P, Vreugde S (2017) Deferiprone and gallium-protoporphyrin have the capacity to potentiate the activity of antibiotics in Staphylococcus aureus small colony variants. Front Cell Infect Microbiol 7:280. https://doi.org/10.3389/fcimb.2017.00280
Sangartit W, Pakdeechote P, Kukongviriyapan V, Donpunha W, Shibahara S, Kukongviriyapan U (2016) Tetrahydrocurcumin in combination with deferiprone attenuates hypertension, vascular dysfunction, baroreflex dysfunction, and oxidative stress in iron-overloaded mice. Vascul Pharmacol 87:199–208. https://doi.org/10.1016/j.vph.2016.10.001
Savage KA, Parquet MC, Allan DS, Davidson RJ, Holbein BE, Lilly EA, Fidel PLJ (2018) Iron restriction to clinical isolates of Candida albicans by the novel chelator DIBI inhibits growth and increases sensitivity to azoles in vitro and in vivo in a murine model of experimental vaginitis. Antimicrob Agents Chemother 62(8):e02576-e12517. https://doi.org/10.1128/AAC.02576-17
Schaible UE, Kaufmann SH (2004) Iron and microbial infection. Nat Rev Microbiol 2(12):946–953. https://doi.org/10.1038/nrmicro1046
Schaible UE, Collins HL, Priem F, Kaufmann SH (2002) Correction of the iron overload defect in beta-2-microglobulin knockout mice by lactoferrin abolishes their increased susceptibility to tuberculosis. J Exp Med 196(11):1507–1513. https://doi.org/10.1084/jem.20020897
Schroeder M, Brooks BD, Brooks AE (2017) The complex relationship between virulence and antibiotic resistance. Genes 8:39. https://doi.org/10.3390/genes8010039
Scott C, Holbein BE, Lehmann C (2020) Iron should be restricted in acute infection. Front Biosci Landmark 25(71):673–682. https://doi.org/10.2741/4827
Sheldon JR, Heinrichs DE (2015) Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiol Rev 39(4):592–630. https://doi.org/10.1093/femsre/fuv009
Sheldon JR, Laakso HA, Heinrichs DE (2016) Iron acquisition strategies of bacterial pathogens. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.VMBF-0010-2015
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss R, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC (2016) The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315(8):801–810. https://doi.org/10.1001/jama.2016.0287
Stojiljkovic I, Kumar V, Srinivasan N (1999) Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol Microbiol 31(2):429–442. https://doi.org/10.1046/j.1365-2958.1999.01175.x
Sukhbaatar N, Weichhart T (2018) Iron regulation: macrophages in control. Pharmaceuticals 11(4):137. https://doi.org/10.3390/ph11040137
Tacke F, Nuraldeen R, Koch A, Strathmann K, Hutschenreuter G, Trautwein C, Strnad P (2016) Iron parameters determine the prognosis of critically ill patients. Crit Care Med 44(6):1049–1058. https://doi.org/10.1097/CCM.0000000000001607
Thompson MG, Corey BW, Si Y, Craft DW, Zurawski DV (2012) Antibacterial activities of iron chelators against common nosocomial pathogens. Antimicrob Agents Chemother 56(10):5419–5421. https://doi.org/10.1128/AAC.01197-12
Thorburn T, Aali M, Kostek L, LeTourneau-Paci C, Colp P, Zhou J, Holbein BE, Hoskin D, Lehmann C (2017) Anti-inflammatory effects of a novel iron chelator, DIBI, in experimental sepsis. Clin Hemorheol Microcirc 67(3–4):241–250. https://doi.org/10.3233/CH-179205
Tolosano E, Altruda F (2002) Hemopexin: structure, function, and regulation. DNA Cell Biol 21(4):297–306. https://doi.org/10.1089/104454902753759717
Vlachodimitropoulou E, Chen Y-L, Garbowski M, Koonyosying P, Psaila B, Sola-Visner M, Cooper N, Hider R, Porter J (2017) Eltrombopag: a powerful chelator of cellular or extracellular iron(III) alone or combined with a second chelator. Blood 130(17):1923–1933. https://doi.org/10.1182/blood-2016-10-740241
Wagener BM, Hu PJ, Oh J-Y, Evans CA, Richter JR, Honavar J, Brandon AP, Creighton J, Stephens SW, Morgan C, Dull RO, Marques MB, Kerby JD, Pittet J-F, Patel RP (2018) Role of heme in lung bacterial infection after trauma hemorrhage and stored red blood cell transfusion: a preclinical experimental study. PLoS Med 15(3):e1002522. https://doi.org/10.1371/journal.pmed.1002522
Ward PP, Uribe-Luna S, Conneely OM (2002) Lactoferrin and host defense. Biochem Cell Biol 80(1):95–102
Weinberg ED (1978) Iron and infection. Microbiol Rev 42(1):45–66
Weinberg ED (2009) Iron availability and infection. Biochem Biophys Acta 1790:600–605. https://doi.org/10.1016/j.bbagen.2008.07.002
Wilson BR, Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y (2016) Siderophores in iron metabolism: from mechanism to therapy potential. Trends Mol Med 22(12):1077–1090. https://doi.org/10.1016/j.molmed.2016.10.005
Wright GD (2005) Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv Drug Deliv Rev 57:1451–1470. https://doi.org/10.1016/j.addr.2005.04.002
Wunderer F, Traeger L, Sigurslid HH, Meybohm P, Bloch DB, Malhotra R (2020) The role of hepcidin and iron homeostasis in atherosclerosis. Pharmacol Res 153:104664. https://doi.org/10.1016/j.phrs.2020.104664
Wyckoff EE, Mey AR, Payne SM (2007) Iron acquisition in Vibrio cholerae. Biometals 20(3):405. https://doi.org/10.1007/s10534-006-9073-4
Zeidan AM, Griffiths EA (2018) To chelate or not to chelate in MDS: that is the question! Blood Rev 32(5):368–377. https://doi.org/10.1016/j.blre.2018.03.002
Zhou T, Ma Y, Kong X, Hider RC (2012) Design of iron chelators with therapeutic application. Dalton Trans 41(21):6371–6389. https://doi.org/10.1039/c2dt12159j
Zhou Y-J, Zhang M-X, Hider RC, Zhou T (2014) In vitro antimicrobial activity of hydroxypyridinone hexadentate-based dendrimeric chelators alone and in combination with norfloxacin. Fed Eur Microbiol Soc Microbiol Lett 355:124–130. https://doi.org/10.1111/1574-6968.12465
Zhou Y-J, Kong X-L, Li J-P, Ma Y-M, Hider R-C, Zhou T (2015a) Novel 3-hydroxypyridin-4-one hexadentate ligand-based polymeric iron chelator: synthesis, characterization and antimicrobial evaluation. Med Chem Commun 6:1620–1625. https://doi.org/10.1039/C5MD00264H
Zhou Y-J, Liu M-S, Osamah AR, Kong X-L, Alsam S, Battah S, Xie Y-Y, Hider RC, Zhou T (2015b) Hexadentate 3-hydroxypyridin-4-ones with high iron(III) affinity: design, synthesis and inhibition on methicillin resistant Staphylococcus aureus and Pseudomonas strains. Eur J Med Chem 94:8–21. https://doi.org/10.1016/j.ejmech.2015.02.050
Zhou T, Chena K, Kong L-M, Liu M-S, Mac Y-M, Xie Y-Y (2018) Synthesis, iron binding and antimicrobial properties of hexadentate 3-hydroxypyridinones-terminated dendrimers. Bioorg Med Chem Lett 28:2504–2512. https://doi.org/10.1016/j.bmcl.2018.05.058
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
BEH has a beneficial interest in Chelation Partners Inc. The other authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Holbein, B.E., Ang, M.T.C., Allan, D.S. et al. Iron-withdrawing anti-infectives for new host-directed therapies based on iron dependence, the Achilles’ heel of antibiotic-resistant microbes. Environ Chem Lett 19, 2789–2808 (2021). https://doi.org/10.1007/s10311-021-01242-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10311-021-01242-7