Abstract
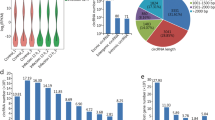
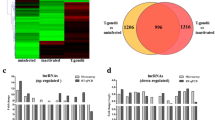
Trichosporon asahii (T. asahii) is a clinically important opportunistic pathogenic fungus capable of causing systemic lethal infection in immunosuppressive and immunodeficient hosts. However, the mechanism of the host immune response upon T. asahii infection has not been elucidated. Recent evidence has shown that long noncoding RNAs (lncRNAs) play key roles in regulating the immune response to resist microbial infections. In this study, we analyzed the expression profiles of lncRNAs at 12 and 24 h post-infection (hpi) in THP-1 cells infected with T. asahii using RNA sequencing (RNA-Seq). A total of 64 and 160 lncRNAs displayed significant differentially expressed (DE) at 12 h and 24 hpi, respectively. Among these lncRNAs, 18 lncRNAs were continuous DE at two time points. The DE of eight candidate lncRNAs were verified by real time quantitative polymerase chain reaction (RT-qPCR). Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses were performed to analyze the cis-target genes of 18 DE lncRNAs. The results showed that they were enriched in signaling pathways related to the host immune response, indicating that these lncRNAs might play important roles in fungi–host interactions. Finally, we explored the function of lncRNA NEAT1 and found that the expression of TNF-α and IL-1β declined after NEAT1 knockdown in T. asahii-infected THP-1 cells. To our knowledge, this is the first report of a expression analysis of lncRNAs in macrophages infected with T. asahii. Our study helps to elucidate the role of lncRNAs in the host immune response to early infection by T. asahii.





Similar content being viewed by others
Data availability
The data used to support the findings of this study are included within the supplementary files.
References
Marine M, Brown NA, Riano-Pachon DM, Goldman GH. On and under the skin: emerging basidiomycetous yeast infections caused by trichosporon species. PLoS Pathog. 2015;11(7):e1004982. https://doi.org/10.1371/journal.ppat.1004982.
Colombo AL, Padovan AC, Chaves GM. Current knowledge of Trichosporon spp. and Trichosporonosis. Clinical Microbiol Rev. 2011;24(4):682–700. https://doi.org/10.1128/CMR.00003-11.
Zhang D, Lu X, Liao Y, Xia Z, Peng Z, Yang X, et al. Rapid and simple detection of by optimized colony PCR. Biomed Res Int. 2019;2019:1803278. https://doi.org/10.1155/2019/1803278.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. https://doi.org/10.1038/nature07672.
Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17(12):756–70. https://doi.org/10.1038/nrm.2016.126.
Lu H, Liu H, Yang X, Ye T, Lv P, Wu X, et al. LncRNA BLACAT1 may serve as a prognostic predictor in cancer: evidence from a meta-analysis. Biomed Res Int. 2019. https://doi.org/10.1155/2019/1275491.
Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19(3):143–57. https://doi.org/10.1038/nrm.2017.104.
Ma YM, Ouyang J, Wei JY, Maarouf M, Chen JL. Involvement of host non-coding RNAs in the pathogenesis of the influenza virus. Int J Mol Sci. 2017;18(1):18. https://doi.org/10.3390/ijms18010039.
Huang S, Huang Z, Luo Q, Qing C. The expression of lncRNA NEAT1 in human tuberculosis and its antituberculosis effect. Biomed Res Int. 2018. https://doi.org/10.1155/2018/9529072.
Wang M, Wang F, Yang J, Zhao D, Wang H, Shao F, et al. Mannan-binding lectin inhibits Candida albicans-induced cellular responses in PMA-activated THP-1 cells through Toll-like receptor 2 and Toll-like receptor 4. PLoS ONE. 2013;8(12):e83517. https://doi.org/10.1371/journal.pone.0083517.
Agustinho DP, de Oliveira MA, Tavares AH, Derengowski L, Stolz V, Guilhelmelli F, et al. Dectin-1 is required for miR155 upregulation in murine macrophages in response to Candida albicans. Virulence. 2017;8(1):41–52. https://doi.org/10.1080/21505594.2016.1200215.
Ganesan S, Rathinam VAK, Bossaller L, Army K, Kaiser WJ, Mocarski ES, et al. Caspase-8 modulates dectin-1 and complement receptor 3-driven IL-1β production in response to β-glucans and the fungal pathogen, Candida albicans. J Immunol (Baltimore Md: 1950). 2014;193(5):2519–30. https://doi.org/10.4049/jimmunol.1400276.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. https://doi.org/10.1093/bioinformatics/btu170.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. https://doi.org/10.1038/nmeth.3317.
Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–5. https://doi.org/10.1093/bioinformatics/bts356.
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–5. https://doi.org/10.1038/nbt.3122.
Li A, Zhang J, Zhou Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014;15:311. https://doi.org/10.1186/1471-2105-15-311.
Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–6. https://doi.org/10.1093/nar/gkx428.
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41(17):e166. https://doi.org/10.1093/nar/gkt646.
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–85. https://doi.org/10.1093/nar/gkv1344.
Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015. https://doi.org/10.1093/nar/gku1063.
Nawrocki EP, Eddy SR. Infernal 11 100-fold faster RNA homology searches. Bioinformatics. 2013;29(22):2933–5. https://doi.org/10.1093/bioinformatics/btt509.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. https://doi.org/10.1093/nar/25.17.3389.
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006. https://doi.org/10.1093/nar/gkj112.
Wu Y, Wei B, Liu H, Li T, Rayner S. MiRPara: a SVM-based software tool for prediction of most probable microRNA coding regions in genome scale sequences. BMC Bioinformatics. 2011;12:107. https://doi.org/10.1186/1471-2105-12-107.
Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–7. https://doi.org/10.1038/nature11508.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. https://doi.org/10.1186/s13059-014-0550-8.
The Gene Ontology C. The gene ontology resource: 20 years and still going strong. Nucleic Acids Res. 2019;47(D1):D330–8. https://doi.org/10.1093/nar/gky1055.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. https://doi.org/10.1093/nar/28.1.27.
Zhang P, Cao L, Zhou R, Yang X, Wu M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019;10(1):1495. https://doi.org/10.1038/s41467-019-09482-6.
Bu D, Luo H, Jiao F, Fang S, Tan C, Liu Z, et al. Evolutionary annotation of conserved long non-coding RNAs in major mammalian species. Sci China Life Sci. 2015;58(8):787–98. https://doi.org/10.1007/s11427-015-4881-9.
Agliano F, Rathinam VA, Medvedev AE, Vanaja SK, Vella AT. Long noncoding RNAs in host-pathogen interactions. Trends Immunol. 2019;40(6):492–510. https://doi.org/10.1016/j.it.2019.04.001.
Zhao P, Liu S, Zhong Z, Jiang T, Weng R, Xie M, et al. Analysis of expression profiles of long noncoding RNAs and mRNAs in brains of mice infected by rabies virus by RNA sequencing. Sci Rep. 2018;8(1):11858. https://doi.org/10.1038/s41598-018-30359-z.
Yi Z, Li J, Gao K, Fu Y. Identifcation of differentially expressed long non-coding RNAs in CD4+ T cells response to latent tuberculosis infection. J Infect. 2014;69(6):558–68. https://doi.org/10.1016/j.jinf.2014.06.016.
Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol. 2019;13(1):46–60. https://doi.org/10.1002/1878-0261.12404.
Ma H, Han P, Ye W, Chen H, Zheng X, Cheng L, et al. The Long noncoding RNA Neat1 exerts antihantaviral effects by Acting as positive feedback for RIG-I signaling. J Virol. 2017;91(9):e02250-e2316. https://doi.org/10.1128/JVI.02250-16.
Acknowledgements
The authors express their gratitude to all of the laboratory members for their assistance.
Funding
This work was supported by the National Natural Science Foundation of China (Grant: 81571972, 82002120 and 82073466) and the National Science Foundation of Beijing, China (Grant: 7202201 and 7212105).
Author information
Authors and Affiliations
Contributions
MZ and ZX conducted the experiments and wrote the manuscript. DZ and XY collected and analyzed the data. RY and JA supervised the project. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors have no conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Handling Editor: Celia Maria de Almeida Soares.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, M., Xia, Z., Zhang, D. et al. Use of RNA Sequencing to Perform Comprehensive Analysis of Long Noncoding RNA Expression Profiles in Macrophages Infected with Trichosporon asahii. Mycopathologia 186, 355–365 (2021). https://doi.org/10.1007/s11046-021-00552-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-021-00552-2