Abstract
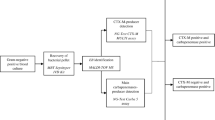
We optimized and prospectively evaluated a simple MALDI-TOF MS-based method for direct detection of third-generation oxymino-cephalosporin resistance (3rd CephR) in Escherichia coli and Klebsiella spp. from blood cultures (BC). In addition, we assessed the performance of a lateral flow immunochromatographic assay (LFIC) for detecting extended-spectrum β-lactamases (ESBL) (NG-Test CTX-M MULTI assay) using bacterial pellets from BC. A total of 168 BCs from unique patients were included. A pre-established volume of BC flagged as positive was transferred in brain heart infusion with or without ceftriaxone (2 mg/ml). After 2-h incubation, intact bacterial pellets were used for MALDI-TOF MS testing. Identification of bacterial species (index score > 2) in the presence of CRO was considered marker of 3rd CephR. The LFIC assay was evaluated in 141 BC. Bacteremia episodes were caused by E. coli (n = 115) or Klebsiella spp. (n = 53). A total of 49 strains were 3rd CephR by broth microdilution, of which 41 were ESBL producers, seven expressed ESBL and OXA-48 type D carbapenemase, and one harbored a plasmid-mediated AmpC. The MALDI-TOF MS method yielded four very major errors (false susceptibility) and two major errors (false resistance). The overall sensitivity of the assay was 91.8% and the specificity 98.3%. Concordance between the LFIC assay and the MALDI-TOF MS method for detection of ESBL-mediated 3rd CephR was 100%. Both evaluated methods may prove useful for early adjustment of empirical therapy in patients with E. coli and Klebsiella spp. bloodstream infections. Whether their use has a beneficial impact on patient outcomes is currently under investigation.

Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH (2000) The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 118:146–155
Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP, Artigas A, Schorr C, Levy MM (2014) Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program. Crit Care Med 42:1749–1755
Kang CI, Kim SH, Park WB, Lee KD, Kim HB, Kim EC, Oh MD, Choe KW (2005) Bloodstream infections caused by antibiotic-resistant Gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother 49:760–766
Shorr AF, Micek ST, Welch EC, Doherty JA, Reichley RM, Kollef MH (2011) Inappropriate antibiotic therapy in Gram-negative sepsis increases hospital length of stay. Crit Care Med 39:46–51
Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ (2020) Infectious Diseases Society of America Antimicrobial Resistant Treatment Guidance: Gram-negative bacterial infections. Clin Infect Dis 27:ciaa1478
Oviaño M, Bou G (2018) Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the rapid detection of antimicrobial resistance mechanisms and beyond. Clin Microbiol Rev 32:e00037–e00018
Faron ML, Buchan BW, Ledeboer NA (2017) Matrix-assisted laser desorption ionization-time of flight mass spectrometry for use with positive blood cultures: methodology, performance, and optimization. J Clin Microbiol 55:3328–3338
Jung JS, Popp C, Sparbier K, Lange C, Kostrzewa M, Schubert S (2014) Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid detection of β-lactam resistance in Enterobacteriaceae derived from blood cultures. J Clin Microbiol 52:924–930
Jung JS, Hamacher C, Gross B, Sparbier K, Lange C, Kostrzewa M, Schubert S (2016) Evaluation of a semiquantitative matrix-assisted laser desorption ionization-time of flight mass spectrometry method for rapid antimicrobial susceptibility testing of positive blood cultures. J Clin Microbiol 54:2820–2824
Sauget M, Bertrand X, Hocquet D (2018) Rapid antibiotic susceptibility testing on blood cultures using MALDI-TOF MS. PLoS One 13:e0205603
Axelsson C, Rehnstam-Holm A-S, Nilso B (2020) Rapid detection of antibiotic resistance in positive blood cultures by MALDI-TOF MS and an automated and optimized MBT-ASTRA protocol for Escherichia coli and Klebsiella pneumoniae. Infect Dis (Lond) 52:45–53
Hernández Egido S, Luis Reboredo A, García Señán A, Gil González AB, Muñoz Bellido JL, González Buitrago JM, Sanchez-Juanes F (2019) Summation of peaks and L34 ribosomal protein in the presence and absence of antibiotics enables susceptibility testing using MALDI-TOF mass spectrometry in 2h from Escherichia coli-positive blood cultures. Enferm Infecc Microbiol Clin 37:244–250
Florio W, Baldeschi L, Rizzato C, Tavanti A, Ghelardi E, Lupetti A (2020) Detection of antibiotic-resistance by MALDI-TOF mass spectrometry: an expanding area. Front Cell Infect Microbiol 10:572909
Torres I, Gimenez E, Pascual T, Bueno F, Huntley D, Martínez M, Navarro D (2017) Short-term incubation of positive blood cultures in brain-heart infusion broth accelerates identification of bacteria by matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry. J Med Microbiol 66:1752–1758
Bevan ER, Jones AM, Hawkey PM (2017) Global epidemiology of CTX-M blactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145e55
Takissian J, Bonnin RA, Naas T, Dortet L (2019) NG-Test Carba 5 for rapid detection of carbapenemase-producing enterobacterales from positive blood cultures. Antimicrob Agents Chemother 63:e00011–e00019
Boutal H, Naas T, Devilliers K, Oueslati S, Dortet L, Bernabeu S, Simon S, Volland H (2017) Development and validation of a lateral flow immunoassay for rapid detection of NDM-producing enterobacteriaceae. J Clin Microbiol 55:2018–2029
Boutal H, Vogel A, Bernabeu S, Devilliers K, Creton E, Cotellon G, Plaisance M, Oueslati S, Dortet L, Jousset A, Oueslati S, Dortet L, Jousset A, Simon S, Naas T, Volland H (2018) A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 73:909–915
Bianco G, Boattini M, Iannaccone M, Cavallo R, Costa C (2020) Evaluation of the NG-Test CTX-M MULTI immunochromatographic assay for the rapid detection of CTX-M extended-spectrum-beta-lactamase producers from positive blood cultures. J Hosp Infect 105:341–343
Bernabeu S, Ratnam KC, Boutal H, Gonzalez C, Vogel A, Devilliers K, Plaisance M, Oueslati S, Malhotra-Kumar S, Dortet L, Fortineau N, Simon S, Volland H, Naas T (2020) A lateral flow immunoassay for the rapid identification of CTX-M-producing enterobacterales from culture plates and positive blood cultures. Diagnostics (Basel) 10:764
Torres Fink I, Tormo Palop N, Borrás Salvador R, Buesa Gómez J, Gimeno Cardona C, Navarro Ortega D (2019) Evaluation of the DNA microarray “AMR Direct Flow Chip Kit” for detection of antimicrobial resistance genes from Gram-positive and Gram-negative bacterial isolated colonies. Enferm Infecc Microbiol Clin 37:454–457
Li M, Liu M, Song Q, Xiong L, Chen Z, Kang M, Xie Y (2018) Rapid antimicrobial susceptibility testing by matrix-assisted laser desorption ionization-time of flight mass spectrometry using a qualitative method in Acinetobacter baumannii complex. J Microbiol Methods 153:60–65
Idelevich EA, Storck LM, Sparbier K, Drews O, Kostrzewa M, Becker K (2018) Rapid direct susceptibility testing from positive blood cultures by the matrix-assisted laser desorption ionization-time of flight mass spectrometry-based direct-on-target microdroplet growth assay. J Clin Microbiol 56:e00913–e00918
Acknowledgements
E.G holds a Juan Rodés research contract (JR18/00053) from the ISCIII (Carlos III Health Institute, Instituto de Salud Carlos III in the original Spanish). E.A. holds a Río Hortega research contract from the Carlos III Health Institute (Ref. CM18/00221).
Author information
Authors and Affiliations
Contributions
IT, EA, EG, JC: conceptualization, methodology, analysis of data, validation, review and editing; BG, AV, TP, DH, DS, RMC: methodology, review and editing; CP, RO: clinical management of patients; DN: conceptualization, supervision, writing the original draft.
Corresponding author
Ethics declarations
Ethical statement
The current study was approved by the Research Ethics Committee of Hospital Clínico Universitario INCLIVA (September, 2019).
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Informed consent
Not applicable (as discussed with the institutional medical ethics committee).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Torres, I., Albert, E., Giménez, E. et al. Performance of a MALDI-TOF mass spectrometry-based method for rapid detection of third-generation oxymino-cephalosporin-resistant Escherichia coli and Klebsiella spp. from blood cultures. Eur J Clin Microbiol Infect Dis 40, 1925–1932 (2021). https://doi.org/10.1007/s10096-021-04251-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-021-04251-0