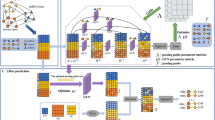
Abstract
The synthetic lethality (SL) relationship arises when a combination of deficiencies in two genes leads to cell death, whereas a deficiency in either one of the two genes does not. The survival of the mutant tumor cells depends on the SL partners of the mutant gene, thereby the cancer cells could be selectively killed by inhibiting the SL partners of the oncogenic genes but normal cells could not. Therefore, there is an urgent need to develop more efficient computational methods of SL pairs identification for cancer targeted therapy. In this paper, we propose a new approach based on similarity fusion to predict SL pairs. Multiple types of gene similarity measures are integrated and k-nearest neighbors algorithm (k-NN) is applied to achieve the similarity-based classification task between gene pairs. As a similarity-based method, our method demonstrated excellent performance in multiple experiments. Besides the effectiveness of our method, the ease of use and expansibility can also make our method more widely used in practice.
Similar content being viewed by others
References
Hartwell L H, Szankasi P, Roberts C J et al. Integrating genetic approaches into the discovery of anticancer drugs. Science, 1997, 278(5340): 1064-1068. https://doi.org/10.1126/science.278.5340.1064.
Boone C, Bussey H, Andrews B J. Exploring genetic interactions and networks with yeast. Nature Reviews Genetics, 2007, 8(6): 437-449. https://doi.org/10.1038/nrg2085.
Chan D A, Giaccia A J. Harnessing synthetic lethal interactions in anticancer drug discovery. Nature Reviews Drug Discovery, 2011, 10(5): 351-364. https://doi.org/10.1038/nrd3374.
Deng X, Das S, Valdez K et al. SL-BioDP: Multi-cancer interactive tool for prediction of synthetic lethality and response to cancer treatment. Cancers (Basel), 2019, 11(11): Article No. 1682. https://doi.org/10.3390/cancers11111682.
McLornan D P, List A, Mufti G J. Applying synthetic lethality for the selective targeting of cancer. New England Journal of Medicine, 2014, 371(18): 1725-1735. https://doi.org/10.1056/NEJMra1407390.
Bryant H E, Schultz N, Thomas H D et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature, 2007, 434(7035): 913-917. https://doi.org/10.1038/nature03443.
Downward J. Targeting RAS signalling pathways in cancer therapy. Nature Reviews Cancer, 2003, 3(1): 11-22. https://doi.org/10.1038/nrc969.
Fong P C, Boss D S, Yap T A et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. New England Journal of Medicine, 2009, 361(2): 123-134. https://doi.org/10.1056/NEJMoa0900212.
Jackson S P, Bartek J. The DNA-damage response in human biology and disease. Nature, 2009, 461(7267): 1071-1078. https://doi.org/10.1038/nature08467.
Lee J S, Das A, Auslander N et al. Harnessing synthetic lethality to predict the response to cancer treatment. Nature Communications, 2018, 9(1): Article No. 2546. https://doi.org/10.1038/s41467-018-04647-1.
Simons A, Dafni N, Dotan I. Establishment of a chemical synthetic lethality screen in cultured human cells. Genome Research, 2001, 11(2): 266-273. https://doi.org/10.1101/gr.154201.
Barbie D A, Tamayo P, Boehm J S et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature, 2009, 462(7269): 108-112. https://doi.org/10.1038/nature08460.
Steckel M, Molina-Arcas M, Weigelt B et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Research, 2012, 22(8): 1227-1245. https://doi.org/10.1038/cr.2012.82.
Han K, Jeng E E, Hess G T et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nature Biotechnology, 2017, 35(5): 463-474. https://doi.org/10.1038/nbt.3834.
Du D, Roguev A, Gordon D E et al. Genetic interaction mapping in mammalian cells using CRISPR interference. Nature Methods, 2017, 14(6): 577-580. https://doi.org/10.1038/nmeth.4286.
Bleicher K H, Böhm H J, Müller K et al. Hit and lead generation: Beyond high-throughput screening. Nature Reviews Drug Discovery, 2003, 2(5): 369-378. https://doi.org/10.1038/nrd1086.
Bajorath J. Integration of virtual and high-throughput screening. Nature Reviews Drug Discovery, 2002, 1(11): 882-894. https://doi.org/10.1038/nrd941.
Guo J, Liu H, Zheng J. SynLethDB: Synthetic lethality database toward discovery of selective and sensitive anticancer drug targets. Nucleic Acids Res., 2016, 44(D1): D1011-D1017. https://doi.org/10.1093/nar/gkv1108.
Lu X, Kensche P R, Huynen M A et al. Genome evolution predicts genetic interactions in protein complexes and reveals cancer drug targets. Nature Communications, 2013, 4: Article No. 2124. https://doi.org/10.1038/ncomms3124.
Srivas R, Shen J P, Yang C C et al. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Molecular Cell, 2016, 63(3): 514-525. https://doi.org/10.1016/j.molcel.2016.06.022.
Kim J W, Botvinnik O B, Abudayyeh O et al. Characterizing genomic alterations in cancer by complementary functional associations. Nature Biotechnology, 2016, 34(5): 539-546. https://doi.org/10.1038/nbt.3527.
Cho H, Berger B, Peng J. Compact integration of multi-network topology for functional analysis of genes. Cell Systems, 2016, 3(6): 540-548. https://doi.org/10.1016/j.cels.2016.10.017.
Jerby-Arnon L, Pfetzer N, Waldman Y et al. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell, 2014, 158(5): 1199-1209. https://doi.org/10.1016/j.cell.2014.07.027.
Wan F, Li S, Tian T et al. EXP2SL: A machine learning framework for cell-line-specific synthetic lethality prediction. Frontiers in Pharmacology, 2020, 11: Article No. 112. https://doi.org/10.3389/fphar.2020.00112.
Liany H, Jeyasekharan A, Rajan V. Predicting synthetic lethal interactions using heterogeneous data sources. Bioinformatics, 2020, 36(7): 2209-2216. https://doi.org/10.1093/bioinformatics/btz893.
Li P, Huang C, Fu Y et al. Large-scale exploration and analysis of drug combinations. Bioinformatics, 2015, 31(12): 2007-2016. https://doi.org/10.1093/bioinformatics/btv080.
Menche J, Sharma A, Kitsak M et al. Uncovering disease-disease relationships through the incomplete interactome. Science, 2015, 347(6224): Article No. 1257601. https://doi.org/10.1126/science.1257601.
Duan Q, Flynn C, Niepel M et al. LINCS Canvas Browser: Interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Research, 2014, 42(W1): W449-W460. https://doi.org/10.1093/nar/gku476.
The UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Research, 2015, 43(D1): D204-D212. https://doi.org/10.1093/nar/gku989.
Davis A P, Grondin C J, Johnson R J et al. The comparative toxicogenomics database: Update 2019. Nucleic Acids Research, 2019, 47(D1): D948-D954. https://doi.org/10.1093/nar/gky868.
Subramanian A, Tamayo P, Mootha V K et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(43): 15545-15550. https://doi.org/10.1073/pnas.0506580102.
Iorio F, Tagliaferri R, Di Bernardo D. Identifying network of drug mode of action by gene expression profiling. Journal of Computational Biology, 2009, 16(2): 241-251. https://doi.org/10.1089/cmb.2008.10TT.
Smith T F, Waterman M S. Identification of common molecular subsequences. Journal of Molecular Biology, 1981, 147(1): 195-197. https://doi.org/10.1016/0022-2836(81)90087-5.
Perlman L, Gottlieb A, Atias N et al. Combining drug and gene similarity measures for drug-target elucidation. Journal of Computational Biology, 2011, 18(2): 133-145. https://doi.org/10.1089/cmb.2010.0213.
Yu G, Li F, Qin Y et al. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics, 2010, 26(7): 976-978. https://doi.org/10.1093/bioinformatics/btq064.
Wang J Z, Du Z, Payattakool R et al. A new method to measure the semantic similarity of GO terms. Bioinformatics, 2007, 23(10): 1274-1281. https://doi.org/10.1093/bioinformatics/btm087.
Wang B, Mezlini A, Demir F et al. Similarity network fusion for aggregating data types on a genomic scale. Nature Methods, 2014, 11(3): 333-337. https://doi.org/10.1038/nmeth.2810.
Altman N S. An introduction to kernel and nearest-neighbor nonparametric regression. The American Statistician, 1992, 46(3): 175-185. https://doi.org/10.1080/00031305.1992.10475879.
He S, He H, Xu W. ICM: A web server for integrated clustering of multi-dimensional biomedical data. Nucleic Acids Research, 2016, 44(W1): W154-W159. https://doi.org/10.1093/nar/gkw378.
Hoadley K A, Yau C, Wolf D M et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell, 2014, 158(4): 929-944. https://doi.org/10.1016/j.cell.2014.06.049.
Ma T, Zhang A. Affinity network fusion and semi-supervised learning for cancer patient clustering. Methods, 2018, 145: 16-24. https://doi.org/10.1016/j.ymeth.2018.05.020.
Tipping M E, Bishop C M. Probabilistic principal component analysis. Journal of the Royal Statistical Society, Series B, 1999, 21(3): 611-622. https://doi.org/10.1111/1467-9868.00196.
Pedregosa F, Varoquaux G, Gramfort A et al. Scikit-learn: Machine learning in Python. Journal of Machine learning Research, 2011, 12: 2825-2830.
Moore A R, Rosenberg S C, McCormick F et al. RAS-targeted therapies: Is the undruggable drugged? Nature Reviews Drug Discovery, 2020, 19(8): 533-552. https://doi.org/10.1038/s41573-020-0068-6.
Wishart D S, Feunang Y D, Guo A C et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research, 2018, 46(D1): D1074-D1082. https://doi.org/10.1093/nar/gkx1037.
Costa-Cabral S, Brough R, Konde A et al. CDK1 is a synthetic lethal target for KRAS mutant tumours. PLoS ONE, 2016, 11(2): Article No. e0149099. https://doi.org/10.1371/journal.pone.0149099.
Grem J L, Voeller D M, Geoffroy F et al. Determinants of trimetrexate lethality in human colon cancer cells. British Journal of Cancer, 1994, 70(6): 1075-1084. https://doi.org/10.1038/bjc.1994.451.
Raimondi M V, Randazzo O, La Franca M et al. DHFR inhibitors: Reading the past for discovering novel anti-cancer agents. Molecules, 2019, 24(6): Article No. 1140. https://doi.org/10.3390/molecules24061140.
Gesto D S, Cerqueira N M, Fernandes P A et al. Gemcitabine: A critical nucleoside for cancer therapy. Current Medicinal Chemistry, 2012, 19(7): 1076-1087. https://doi.org/10.2174/092986712799320682.
Shimasaki T, Ishigaki Y, Nakamura Y et al. Glycogen synthase kinase 3β inhibition sensitizes pancreatic cancer cells to gemcitabine. Journal of Gastroenterology, 2012, 47(3): 321-333. https://doi.org/10.1007/s00535-011-0484-9.
Kunnumakkara A B, Sung B, Ravindran J et al. Zyflamend suppresses growth and sensitizes human pancreatic tumors to gemcitabine in an orthotopic mouse model through modulation of multiple targets. International Journal of Cancer, 2012, 131(3): E292-E303. https://doi.org/10.1002/ijc.26442.
Xia G, Wang H, Song Z et al. Gambogic acid sensitizes gemcitabine efficacy in pancreatic cancer by reducing the expression of ribonucleotide reductase subunit-M2 (RRM2). Journal of Experimental & Clinical Cancer Research, 2017, 36(1): Article No. 107. https://doi.org/10.1186/s13046-017-0579-0.
Yoshida K, Toden S, Ravindranathan P et al. Curcumin sensitizes pancreatic cancer cells to gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1 expression. Carcinogenesis, 2017, 38(10): 1036-1046. https://doi.org/10.1093/carcin/bgx065.
Ashworth A, Lord C J, Reis-Filho J S. Genetic interactions in cancer progression and treatment. Cell., 2011, 145(1): 30-38. https://doi.org/10.1016/j.cell.2011.03.020.
Brough R, Frankum J R, Costa-Cabral S et al. Searching for synthetic lethality in cancer. Current Opinion in Genetics and Development, 2011, 21(1): 34-41. https://doi.org/10.1016/j.gde.2010.10.009.
Author information
Authors and Affiliations
Corresponding authors
Supplementary Information
ESM 1
(PDF 127 kb)
Rights and permissions
About this article
Cite this article
Wu, LL., Wen, YQ., Yang, XX. et al. Synthetic Lethal Interactions Prediction Based on Multiple Similarity Measures Fusion. J. Comput. Sci. Technol. 36, 261–275 (2021). https://doi.org/10.1007/s11390-021-0866-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11390-021-0866-2