Abstract
Cancer is one of the deadly diseases prevailing worldwide and the patients with cancer are rescued only when the cancer is detected at the very early stage. Early detection of cancer is essential as, in the final stage, the chance of survival is limited. The symptoms of cancers are rigorous and therefore, all the symptoms should be studied properly before the diagnosis. Thus, an automatic prediction system is necessary for classifying cancer as malignant or benign. Hence, this paper introduces the novel strategy based on the JayaAnt lion optimization-based Deep recurrent neural network (JayaALO-based DeepRNN) for cancer classification. The steps followed in the developed model are data normalization, data transformation, feature dimension detection, and classification. The first step is data normalization. The goal of data normalization is to eliminate data redundancy and to mitigate the storage of objects in a relational database that maintains the same information in several places. After that, the data transformation is carried out based on log transformation that generates the patterns using more interpretable and helps fulfill the supposition, and to reduce skew. Also, the non-negative matrix factorization is employed for reducing the feature dimension. Finally, the proposed JayaALO-based DeepRNN method effectively classifies cancer based on the reduced dimension features to produce a satisfactory result. Thus, the resulted output of the proposed JayaALO-based DeepRNN is employed for cancer classification. The proposed JayaALO-based DeepRNN showed improved results with maximal accuracy of 95.97%, maximal sensitivity of 95.95%, and maximal specificity of 96.96%.
Graphical abstract

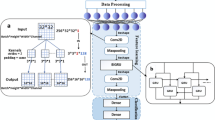
The goal of this research is to devise the cancer classification strategy using the proposed JayaALO-based DeepRNN. It is required to detect the cancer at an early stage to prevent the destruction caused to the other organs. The developed model involves four phases to perform the cancer classification, namely data normalization, data transformation, feature dimension detection, and the classification. Initially, the input images are gathered and are adapted to perform data normalization. The normalized data is fed to the data transformation, which will be performed using log transformation. The obtained transformed data is fed to feature dimension reduction which is performed using non-negative matrix factorization. The reduced features will be employed in DeepRNN for cancer classification. The training of DeepRNN is done using the proposed JayaALO, which is designed by combining ALO and the Jaya algorithm the block diagram of the proposed cancer classification approach using JayaALO-based DeepRNN approach is given below.






Similar content being viewed by others
References
Setti CT, Vogelstein B (2015) Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Br Dent J 347(78):78–81
Scitable by Nature Education, Gene expression. https://www.nature.com/scitable/topicpage/gene-expression. Accessed March 2020
Carpten JC, Mardis ER (2018) The era of precision oncogenomics. Article from Cold Spring Harbor Molecular Case Studies vol. 8
Kourou K, Rigas G, Papaloukas C, Mitsis M, Fotiadis DI (2020) Cancer classification from time series microarray data through regulatory dynamic Bayesian networks. Comput Biol Med 116
Wang SL, Li X, Zhang S, Gui J, Huang DS (2010) Tumor classification by combining PNN classier ensemble with neighborhood rough set based gene reduction. Comput Biol Med 40(2):179–189
Fusco P, Cofini V, Petrucci E, Scimia P, Paladini G, Behr AU, Gobbi F, Pozone T, Danelli G, Di Marco M, Vicentini R, Necozione S, Marinangeli F (2016) Unilateral paravertebral block compared with subarachnoid anesthesia for the management of postoperative pain syndrome after inguinal herniorrhaphy: a randomized controlled. Pain 157(5)
Ma B, Meng F, Yan G, Yan H, Song F (2020) Diagnostic classification of cancers using extreme gradient boosting algorithm and multi-omics data. Comput Biol Med 121
Liu Y (2004) Active learning with support vector machine applied to gene expression data for cancer classification. J Chem Inf Comput Sci 44(6):1936–1941
Tan AC, Gilbert D (2003) Ensemble machine learning on gene expression data for cancer classification. Appl Bioinformatics:S75–S83
Shaik JB, Ganesh V (2020) Deep neural network and social ski-driver optimization algorithm for power system restoration with VSC - HVDC technology. J Comput Mech Power Syst Control 3(1):1–9
Lee H, Grosse R, Ranganath R, Ng AY (2009) Convolutional deep belief networks for scalable unsupervised learning of hierarchical representations. Proceedings of the 26th Annual International Conference on Machine Learning
Danaee P, Ghaeini R, Hendrix DA (2017) A deep learning approach for cancer detection and relevant gene identification. In: Pacific symposium on Biocomputing, pp. 219–229
Brezocnik L, Fister I Jr, Podgorelec V (2018) Swarm intelligence algorithms for feature selection:a review. Appl Sci 8(9):1–31
Snousy MBA, El-Deeb HM, Badran K, Khlil IAA (2011) Suite of decision tree-based classification algorithms on cancer gene expression data. Egypt Informat J 12(2):73–82
Sevakula RK, Singh V, Verma NK, Kumar C, Cui Y (2018) Transfer learning for molecular cancer classification using deep neural networks. IEEE/ACM Trans Comput Biol Bioinform 16(6):2089–2100
Chen Y (2020) Crowd behaviour recognition using Enhanced Butterfly Optimization Algorithm based Recurrent Neural Network. Multimedia Res 3(3)
Bhagyalakshmi V, Ramchandra, Geeta D (2018) Arrhythmia classification using cat swarm optimization based support vector neural network. J Netw Commun Syst 1(1):28–35
Helman P, Veroff R, Atlas SR, Willman C (2004) A Bayesian network classification methodology for gene expression data. J Comput Biol J Comput Mol Cell Biol 11(4):581–615
Halder A, Dey S, Kumar A (2015) Active learning using fuzzy k-NN for cancer classification from microarray gene expression data. Springer, India
Bharathi A, Natarajan AM (2010) Cancer classification of bioinformatics data using ANOVA. Cell Mol Life Sci 2(3):369–373
Shukla AK, Singh P, Vardhan M (2020) Gene selection for cancer types classification using novel hybrid metaheuristics approach. Swarm Evol Comput 54:100661
Sah A, Choudhury T, Rawat S, Tripathi A (2020) A proposed gene selection approach for disease detection. In: Computational intelligence in pattern recognition. Springer, pp 199–206
Liao Q, Ding Y, Jiang ZL, Wang X, Zhang C, Zhang Q (2019) Multi-task deep convolutional neural network for cancer diagnosis. Neurocomputing 348:66–73
Lu H, Gao H, Ye M, Wang X (2019) A hybrid ensemble algorithm combining AdaBoost and genetic algorithm for cancer classification with gene expression data. IEEE/ACM Trans Comput Biol Bioinform
Xu J, Wu P, Chen Y, Meng Q, Dawood H, Khan MM (2019) A novel deep flexible neural forest model for classification of cancer subtypes based on gene expression data. IEEE Access 7:22086–22095
Chaudhari P, Agarwal H, Bhateja V (2019) Data augmentation for cancer classification in oncogenomics: an improved KNN based approach. Evol Intel:1–10
Mudiyanselage TKB, Xiao X, Zhang Y, Pan Y (2019) Deep fuzzy neural networks for biomarker selection for accurate cancer detection. IEEE Trans Fuzzy Syst
Dinakara Prasad Reddy P, Veera Reddy VC, Gowri MT (2018) Ant lion optimization algorithm for optimal sizing of renewable energy resources for loss reduction in distribution systems. J Electr Syst Inf Technol 5(3):663–680
Venkata Rao R (2016) Jaya: a simple and new optimization algorithm for solving constrained and unconstrained optimization problems. Int J Ind Eng Comput 7(1):19–34
Herrero J, Diaz-Uriarte R, Dopazo J (2003) Gene expression data preprocessing. Bioinf Appl Note 19(5):655–656
Hoyer PO (2004) Non-negative matrix factorization with sparseness constraints. J Mach Learn Res 5:1457–1469
Inoue M, Inoue S, Nishida T (2018) Deep recurrent neural network for mobile human activity recognition with high throughput. Artif Life Robot 23(2):173–185
AP_Colon_Kidney dataset taken from https://www.openml.org/d/1137. Accessed April 2020
AP_Breast_Ovary dataset taken from https://www.openml.org/d/1165. Accessed on April 2020
AP_Breast_Colon dataset taken from https://www.openml.org/d/1145. Accessed on April 2020
AP_Breast_Kidney dataset taken from https://www.openml.org/d/1158. Accessed on April 2020
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Majji, R., Nalinipriya, G., Vidyadhari, C. et al. Jaya Ant lion optimization-driven Deep recurrent neural network for cancer classification using gene expression data. Med Biol Eng Comput 59, 1005–1021 (2021). https://doi.org/10.1007/s11517-021-02350-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-021-02350-w