Abstract
Background
The epidermal growth factor receptor (EGFR) is a key protein involved in cancer development. Monoclonal antibodies targeting EGFR are approved for the treatment of metastatic colorectal cancer (CRC). Despite the beneficial clinical effects observed in subgroups of patients, the acquisition of resistance to treatment remains a major concern. Protein N-glycosylation of cellular receptors is known to regulate physiological processes leading to activation of downstream signaling pathways. In the present study, the role of EGFR-specific terminal ⍺2,6-sialylation was analyzed in modulation of the malignant phenotype of CRC cells and their resistance to monoclonal antibody Cetuximab-based therapy.
Methods
Glycoengineered CRC cell models with specific sialyltransferase ST6GAL1 expression levels were applied to evaluate EGFR activation, cell surface glycosylation and therapeutic response to Cetuximab.
Results
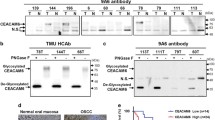
Glycoproteomic analysis revealed EGFR as a major target of ST6Gal1-mediated ⍺2,6-sialylation in a glycosite-specific manner. Mechanistically, CRC cells with increased ST6Gal1 expression and displaying terminal ⍺2,6-sialylation showed a marked resistance to Cetuximab-induced cytotoxicity. Moreover, we found that this resistance was accompanied by downregulation of EGFR expression and its activation.
Conclusions
Our data indicate that EGFR ⍺2,6-sialylation is a key factor in modulating the susceptibility of CRC cells to antibody targeted therapy, thereby disclosing a potential novel biomarker and providing key molecular information for tailor made anti-cancer strategies.





Similar content being viewed by others
Data availability
The mass spectrometry proteomic data supporting the conclusions of this article are available in the ProteomeXchange Consortium via the PRIDE partner repository, under the project name “EGFR N-Glycosylation Profile from Colorectal Cancer Cells”, with the dataset identifier PXD017914.
Code availability
Not applicable.
Abbreviations
- ⍺2,3NeuAc:
-
⍺2,3-linked sialic acid
- ⍺2,6NeuAc:
-
⍺2,6-linked sialic acid
- AAL:
-
Aleuria aurantia lectin
- BrdU:
-
bromodeoxyuridine
- CRC:
-
colorectal cancer
- CRISPR/Cas9:
-
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9
- ECL:
-
enhanced chemiluminescence
- EGFR:
-
epidermal growth factor receptor
- FBS:
-
fetal bovine serum
- gRNA:
-
guide RNA
- IDAA:
-
Indel Detection by Amplicon Analysis
- Indel:
-
insertions and deletions
- KO:
-
Knock-Out
- LTL:
-
Lotus tetragonolobus lectin
- mAb:
-
monoclonal antibody
- MAL-I:
-
Maackia amurensis lectin I
- MFI:
-
median fluorescence intensity
- OE:
-
Overexpression
- ON:
-
overnight
- RT:
-
room temperature
- RTK:
-
receptor tyrosine kinase
- SNA:
-
Sambucus nigra agglutinin
- ST6Gal1:
-
β-Galactoside ⍺2,6-sialyltransferase 1
- TIDE:
-
Tracking of Indels by DEcomposition
- TNFR1:
-
tumor necrosis receptor 1
- UEA-I:
-
Ulex europaeus agglutinin I
- WT:
-
wild-type
References
F. Bray, J. Ferlay, I. Soerjomataram, R.L. Siegel, L.A. Torre, A. Jemal, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 68, 394–424 (2018)
E. Van Cutsem, A. Cervantes, R. Adam, A. Sobrero, J.H. Van Krieken, D. Aderka, E. Aranda Aguilar, A. Bardelli, A. Benson, G. Bodoky, F. Ciardiello, A. D’Hoore, E. Diaz-Rubio, J.Y. Douillard, M. Ducreux, A. Falcone, A. Grothey, T. Gruenberger, K. Haustermans, V. Heinemann, P. Hoff, C.H. Köhne, R. Labianca, P. Laurent-Puig, B. Ma, T. Maughan, K. Muro, N. Normanno, P. Österlund, W.J.G. Oyen, D. Papamichael, G. Pentheroudakis, P. Pfeiffer, T.J. Price, C. Punt, J. Ricke, A. Roth, R. Salazar, W. Scheithauer, H.J. Schmoll, J. Tabernero, J. Taïeb, S. Tejpar, H. Wasan, T. Yoshino, A. Zaanan, D. Arnold, ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 27, 1386–1422 (2016)
D.J. Jonker, C.J. O’Callaghan, C.S. Karapetis, J.R. Zalcberg, D. Tu, H.-J. Au, S.R. Berry, M. Krahn, T. Price, R.J. Simes, N.C. Tebbutt, G. van Hazel, R. Wierzbicki, C. Langer, M.J. Moore, Cetuximab for the treatment of colorectal cancer. N. Engl. J. Med. 357, 2040–2048 (2007)
E.V. Cutsem, M. Peeters, S. Siena, Y. Humblet, A. Hendlisz, B. Neyns, J.-L. Canon, J.-L.V. Laethem, J. Maurel, G. Richardson, M. Wolf, R.G. Amado, Open-label phase iii trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J. Clin. Oncol. 25, 1658–1664 (2007)
S. Li, K.R. Schmitz, P.D. Jeffrey, J.J.W. Wiltzius, P. Kussie, K.M. Ferguson, Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 7, 301–311 (2005)
C.S. Karapetis, S. Khambata-Ford, D.J. Jonker, C.J. O’Callaghan, D. Tu, N.C. Tebbutt, R.J. Simes, H. Chalchal, J.D. Shapiro, S. Robitaille, T.J. Price, L. Shepherd, H.-J. Au, C. Langer, M.J. Moore, J.R. Zalcberg, K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 359, 1757–1765 (2008)
R.G. Amado, M. Wolf, M. Peeters, E.V. Cutsem, S. Siena, D.J. Freeman, T. Juan, R. Sikorski, S. Suggs, R. Radinsky, S.D. Patterson, D.D. Chang, Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 26, 1626–1634 (2008)
A. Bardelli, S. Siena, Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J. Clin. Oncol. 28, 1254–1261 (2010)
A. Varki, Biological roles of glycans. Glycobiology 27, 3–49 (2017)
S.S. Pinho, C.A. Reis, Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 (2015)
S. Mereiter, M. Balmaña, D. Campos, J. Gomes, C.A. Reis, Glycosylation in the era of cancer-targeted therapy: where are we heading? Cancer Cell 36, 6–16 (2019)
S. Holst, M. Wuhrer, Y. Rombouts, Glycosylation characteristics of colorectal cancer. Adv. Cancer Res. 126, 203–256 (2015)
A.S. Carvalho, A. Harduin-Lepers, A. Magalhães, E. Machado, N. Mendes, L.T. Costa, R. Matthiesen, R. Almeida, J. Costa, C.A. Reis, Differential expression of α-2,3-sialyltransferases and α-1,3/4-fucosyltransferases regulates the levels of sialyl Lewis a and sialyl Lewis x in gastrointestinal carcinoma cells. Int. J. Biochem. Cell Biol. 42, 80–89 (2010)
N.T. Marcos, E.P. Bennett, J. Gomes, A. Magalhae, C. Gomes, L. David, I. Dar, C. Jeanneau, S. DeFrees, D. Krustrup, L.K. Vogel, E.H. Kure, J. Burchell, J. Taylor-Papadimitriou, H. Clausen, U. Mandel, C.A. Reis, ST6GalNAc-I controls expression of sialyl-Tn antigen in gastrointestinal tissues. Front. Biosci. 3, 1443-1455 (2011)
F. Dall’Olio, N. Malagolini, M. Trinchera, M. Chiricolo, Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. BBA. Gen. Subj. 1840, 2752–2764 (2014)
J. Weinstein, E.U. Lee, K. McEntee, P.H. Lai, J.C. Paulson, Primary structure of beta-galactoside alpha 2,6-sialyltransferase. Conversion of membrane-bound enzyme to soluble forms by cleavage of the NH2-terminal signal anchor. J. Biol. Chem. 262, 17735–17743 (1987)
F. Dall’Olio, The sialyl-α2,6-lactosaminyl-structure: Biosynthesis and functional role. Glycoconj. J. 17, 669–676 (2000)
S. Zhang, J. Lu, Z. Xu, X. Zou, X. Sun, Y. Xu, A. Shan, J. Lu, X. Yan, Y. Cui, W. Yan, Y. Du, J. Gu, M. Zheng, B. Feng, Y. Zhang, Differential expression of ST6GAL1 in the tumor progression of colorectal cancer. Biochem. Biophys. Res. Commun. 486, 1090–1096 (2017)
P. Geßner, S. Riedl, A. Quentmaier, W. Kemmner, Enhanced activity of CMP-NeuAc:Galβ1-4GlcNAc:α2,6-sialyltransferase in metastasizing human colorectal tumor tissue and serum of tumor patients. Cancer Lett. 75, 143–149 (1993)
C. Costa-Nogueira, S. Villar-Portela, E. Cuevas, E. Gil-Martín, Fernández-Briera, Synthesis and expression of CDw75 antigen in human colorectal cancer. BMC Cancer 9, 431 (2009)
J.-J. Park, M. Lee, Increasing the α 2,6 sialylation of glycoproteins may contribute to metastatic spread and therapeutic resistance in colorectal cancer. Gut Liver 7, 629–641 (2013)
J. Lu, T. Isaji, S. Im, T. Fukuda, N. Hashii, D. Takakura, N. Kawasaki, J. Gu, β-Galactoside α2,6-sialyltranferase 1 promotes transforming growth factor-β-mediated epithelial-mesenchymal transition. J. Biol. Chem. 289, 34627–34641 (2014)
C.M. Britain, N. Bhalerao, A.D. Silva, A. Chakraborty, D.J. Buchsbaum, M.R. Crowley, D.K. Crossman, Y.J.K. Edwards, S.L. Bellis, Glycosyltransferase ST6Gal-I promotes the epithelial to mesenchymal transition in pancreatic cancer cells. J. Biol. Chem. 296, 100034 (2020)
Y. Zhuo, S.L. Bellis, Emerging role of α2,6-sialic acid as a negative regulator of galectin binding and function. J. Biol. Chem. 286, 5935–5941 (2011)
D.O. Croci, J.P. Cerliani, T. Dalotto-Moreno, S.P. Méndez-Huergo, I.D. Mascanfroni, S. Dergan-Dylon, M.A. Toscano, J.J. Caramelo, J.J. García-Vallejo, J. Ouyang, E.A. Mesri, M.R. Junttila, C. Bais, M.A. Shipp, M. Salatino, G.A. Rabinovich, Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell 156, 744–758 (2014)
Y. Zhen, R.M. Caprioli, J.V. Staros, Characterization of glycosylation sites of the epidermal growth factor receptor. Biochemistry 42, 5478–5492 (2003)
C. Sato, J.-H. Kim, Y. Abe, K. Saito, S. Yokoyama, D. Kohda, Characterization of the N-oligosaccharides attached to the atypical Asn-X-Cys sequence of recombinant human epidermal growth factor receptor. J. Biochem. 127, 65–72 (2000)
H.-B. Guo, H. Johnson, M. Randolph, I. Lee, M. Pierce, Knockdown of GnT-Va expression inhibits ligand-induced downregulation of the epidermal growth factor receptor and intracellular signaling by inhibiting receptor endocytosis. Glycobiology 19, 547–559 (2009)
Y. Sato, M. Takahashi, Y. Shibukawa, S.K. Jain, R. Hamaoka, J. Miyagawa, Y. Yaginuma, K. Honke, M. Ishikawa, N. Taniguchi, Overexpression of N-acetylglucosaminyltransferase III enhances the epidermal growth factor-induced phosphorylation of ERK in HeLaS3 cells by up-regulation of the internalization rate of the receptors. J. Biol. Chem. 276, 11956–11962 (2001)
Y.-C. Liu, H.-Y. Yen, C.-Y. Chen, C.-H. Chen, P.-F. Cheng, Y.-H. Juan, C.-H. Chen, K.-H. Khoo, C.-J. Yu, P.-C. Yang, T.-L. Hsu, C.-H. Wong, Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc. Natl. Acad. Sci. U. S. A. 108, 11332–11337 (2011)
J.-J. Park, J.Y. Yi, Y.B. Jin, Y.-J. Lee, J.-S. Lee, Y.-S. Lee, Y.-G. Ko, M. Lee, Sialylation of epidermal growth factor receptor regulates receptor activity and chemosensitivity to gefitinib in colon cancer cells. Biochem. Pharmacol. 83, 849–857 (2012)
Y. Narimatsu, H.J. Joshi, Z. Yang, C. Gomes, Y.-H. Chen, F.C. Lorenzetti, S. Furukawa, K.T. Schjoldager, L. Hansen, H. Clausen, E.P. Bennett, H.H. Wandall, A validated gRNA library for CRISPR/Cas9 targeting of the human glycosyltransferase genome. Glycobiology 28, 295–305 (2018)
E.K. Brinkman, T. Chen, M. Amendola, B. van Steensel, Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014)
M. Bern, Y.J. Kil, C. Becker, Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics. 40, 13.20.11–13.20.14 (2012)
D. Ahmed, P.W. Eide, I.A. Eilertsen, S.A. Danielsen, M. Eknæs, M. Hektoen, G.E. Lind, R.A. Lothe, Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2, e71 (2013)
J.N. Contessa, M.S. Bhojani, H.H. Freeze, A. Rehemtulla, T.S. Lawrence, Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells. Cancer Res. 68, 3803–3809 (2008)
S. Holst, A.J.M. Deuss, G.W. van Pelt, S.J. van Vliet, J.J. Garcia-Vallejo, C.A.M. Koeleman, A.M. Deelder, W.E. Mesker, R.A. Tollenaar, Y. Rombouts, M. Wuhrer, N-glycosylation profiling of colorectal cancer cell lines reveals association of Fucosylation with differentiation and caudal type Homebox 1 (CDX1)/Villin mRNA expression. Mol. Cell. Proteomics 15, 124–140 (2016)
N. Very, T. Lefebvre, I.El Yazidi-Belkoura, Drug resistance related to aberrant glycosylation in colorectal cancer. Oncotarget 9, 1380–1402 (2017)
J.G. Rodrigues, M. Balmaña, J.A. Macedo, J. Poças, Â Fernandes, J.C.M. de-Freitas-Junior, S.S. Pinho, J. Gomes, A. Magalhães, C. Gomes, S. Mereiter, C.A. Reis, Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cell. Immunol. 333, 46–57 (2018)
K. Kaszuba, M. Grzybek, A. Orłowski, R. Danne, T. Róg, K. Simons, Ü Coskun, I. Vattulainen, N-Glycosylation as determinant of epidermal growth factor receptor conformation in membranes. Proc. Natl. Acad. Sci. U. S. A. 112, 4334–4339 (2015)
J. Lu, J. Gu, Significance of β-galactoside α2,6 sialyltranferase 1 in cancers. Molecules 20, 7509–7527 (2015)
M.J. Schultz, A.F. Swindall, J.W. Wright, E.S. Sztul, C.N. Landen, S.L. Bellis, ST6Gal-I sialyltransferase confers cisplatin resistance in ovarian tumor cells. J. Ovarian Res. 6, 25 (2013)
P.R. Punch, E.E. Irons, C.T. Manhardt, H. Marathe, J.T.Y. Lau, The sialyltransferase ST6GAL1 protects against radiation-induced gastrointestinal damage. Glycobiology 30, 446-453 (2020)
C.M. Britain, K.A. Dorsett, S.L. Bellis, The glycosyltransferase ST6Gal-I protects tumor cells against serum growth factor withdrawal by enhancing survival signaling and proliferative potential. J. Biol. Chem. 292, 4663–4673 (2017)
R.B. Jones, K.A. Dorsett, A.B. Hjelmeland, S.L. Bellis, The ST6Gal-I sialyltransferase protects tumor cells against hypoxia by enhancing HIF-1α signaling. J. Biol. Chem. 293, 5659–5667 (2018)
M.J. Schultz, A.T. Holdbrooks, A. Chakraborty, W.E. Grizzle, C.N. Landen, D.J. Buchsbaum, M.G. Conner, R.C. Arend, K.J. Yoon, C.A. Klug, D.C. Bullard, R.A. Kesterson, P.G. Oliver, A.K. O’Connor, B.K. Yoder, S.L. Bellis, The tumor-associated glycosyltransferase ST6Gal-I regulates stem cell transcription factors and confers a cancer stem cell phenotype. Cancer Res. 76, 3978–3988 (2016)
A.F. Swindall, A.I. Londoño-Joshi, M.J. Schultz, N. Fineberg, D.J. Buchsbaum, S.L. Bellis, ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer Res. 73, 2368–2378 (2013)
E.C. Seales, G.A. Jurado, B.A. Brunson, J.K. Wakefield, A.R. Frost, S.L. Bellis, Hypersialylation of β1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 65, 4645–4652 (2005)
A.F. Swindall, S.L. Bellis, Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J. Biol. Chem. 286, 22982–22990 (2011)
Q. Zhang, J.N. Higginbotham, D.K. Jeppesen, Y.-P. Yang, W. Li, E.T. McKinley, R. Graves-Deal, J. Ping, C.M. Britain, K.A. Dorsett, C.L. Hartman, D.A. Ford, R.M. Allen, K.C. Vickers, Q. Liu, J.L. Franklin, S.L. Bellis, R.J. Coffey, Transfer of functional cargo in exomeres. Cell Rep. 27, 940-954.e6 (2019)
H. Zhang, D. Freitas, H.S. Kim, K. Fabijanic, Z. Li, H. Chen, M.T. Mark, H. Molina, A.B. Martin, L. Bojmar, J. Fang, S. Rampersaud, A. Hoshino, I. Matei, C.M. Kenific, M. Nakajima, A.P. Mutvei, P. Sansone, W. Buehring, H. Wang, J.P. Jimenez, L. Cohen-Gould, N. Paknejad, M. Brendel, K. Manova-Todorova, A. Magalhães, J.A. Ferreira, H. Osório, A.M. Silva, A. Massey, J.R. Cubillos-Ruiz, G. Galletti, P. Giannakakou, A.M. Cuervo, J. Blenis, R. Schwartz, M.S. Brady, H. Peinado, J. Bromberg, H. Matsui, C.A. Reis, D. Lyden, Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 20, 332–343 (2018)
M.E. Irwin, K.L. Mueller, N. Bohin, Y. Ge, J.L. Boerner, Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J. Cell. Physiol. 226, 2316–2328 (2011)
M.P. Mathew, E. Tan, C.T. Saeui, P. Bovonratwet, S. Sklar, R. Bhattacharya, K.J. Yarema, Metabolic flux-driven sialylation alters internalization, recycling, and drug sensitivity of the epidermal growth factor receptor (EGFR) in SW1990 pancreatic cancer cells. Oncotarget 7, 66491–66511 (2016)
Acknowledgements
The authors acknowledge Merck KGaA, Darmstadt, Germany for providing the anti-Cetuximab and anti-Matuzumab antibodies. The authors acknowledge Dr. Anne Harduin-Lepers and Dr. Virginie Cogez for their help with the preparation of the pcDNA3.1/Hygro(+)ST6Gal1 plasmid. The authors acknowledge Dr. Luis Cirnes from Ipatimup Diagnostics for the support on the analysis of cell line genetic statuses. The authors acknowledge the support of the i3S Advanced Light Microscopy facility, member of the national infrastructure PPBI - Portuguese Platform of Bioimaging (PPBI-POCI-01-0145-FEDER-022122) and the i3S Proteomics Scientific Platform, member of the Portuguese Mass Spectrometry Network, integrated in the National Roadmap of Research Infrastructures of Strategic Relevance (ROTEIRO/0028/2013; LISBOA-01-0145-FEDER-0221 25). The authors acknowledge the i3S Translational Research and Industry Partnership Office for all the support given for the scientific design and for the continuous monitoring of the project.
Funding
This research was funded by Merck KGaA, Darmstadt, Germany, and by FEDER funds through the Operational Programme for Competitiveness Factors COMPETE 2020 (POCI-01-0145-FEDER-016585; POCI-01-0145-FEDER-007274) and national funds through the Foundation for Science and Technology (FCT), under the projects: PTDC/BBB-EBI/0567/2014 to C.A.R and UID/BIM/04293/2013; PTDC/MED-QUI/29,780/2017 to C.G., and the project NORTE-01-0145-FEDER-000029, supported by Norte Portugal Regional Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF). J.G.R. was supported by a FCT PhD grant (SFRH/BD/136,736/2018); H.O.D. was supported by a FCT PhD grant (PD/BD/128,407/2017) through the FCT PhD Programmes and by Programa Operacional Potencial Humano (POPH), specifically by the BiotechHealth Programme (Doctoral Programme on Cellular and Molecular Biotechnology Applied to Health Sciences), with the reference PD/0016/2012 funded by FCT. M.B. was supported by the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement n.º 748,880.
Author information
Authors and Affiliations
Contributions
J.G. and C.A.R. designed and supervised the study. J.G.R., H.O.D., C.G., M.B., A.M., A.H.d.R. and J.G performed experiments; J.G.R., H.O.D., C.G., P.J.H., J.L., A.A., P.A.V., M.W., J.G. and C.A.R performed the formal analyses; J.G.R., H.O.D., J.G. and C.A.R wrote the original manuscript. All authors reviewed and edited the manuscript. All authors have read and agreed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
Fig. S1 Genomic validation of CRISPR/Cas9 ST6GAL1 KO in Caco-2 cell line. a Validation of nucleotide insertions and deletions (indels) at the ST6GAL1 locus following genome editing by CRISPR/Cas9, in three isogenic Caco-2 ST6Gal1 KO cell clones (KO 17, KO 33 and KO 42) by Indel Detection by Amplicon Analysis (IDAA) PCR. b Validation of Caco-2 ST6Gal1 KO cell clones indels using the Tracking of Indels by DEcomposition (TIDE) bioinformatic tool. Fig. S2 ST6Gal1 overexpression induces the enrichment of EGFR N444 and N530 glycosites in terminally sialylated species. a Upper panel - higher energy collision dissociation (HCD) MS/MS spectra of the N444 glycopeptide modified with a mono-sialylated biantennary N-glycan in SW48 ST6Gal1 OE 1 EGFR; Middle panel – collision-induced dissociation (CID) MS/MS spectrum of the same glycopeptide; Bottom panel – HCD MS/MS spectra of the same glycopeptide depicting identified b and y ions for peptide identification. b Upper panel - HCD MS/MS spectra of the N528 glycopeptide modified with a di-sialylated biantennary N-glycan in SW48 ST6Gal1 OE 1 EGFR; Middle panel – CID MS/MS spectrum of the same glycopeptide; Bottom panel – HCD MS/MS spectra of the same glycopeptide depicting identified b and y ions for peptide identification. Fig. S3 EGFR site-specific glycan composition in Caco-2 cells. a Detection of ⍺2,6-linked sialic acid (⍺2,6NeuAc) structures in immunoprecipitated EGFR from Caco-2 WT and ST6Gal1 KO (KO 17, KO 33 and KO 42) cell clones by reactivity with SNA. b Colloidal Blue gel staining of immunoprecipitated EGFR from Caco-2 WT and ST6Gal1 KO cell clones. c Schematic representation of EGFR glycosylation following site-specific assignment and structural glycan characterization in Caco-2 WT and ST6Gal1 KO cell clones. Fig. S4. Impact of ST6Gal1 overexpression in SW48 cells proliferation, cell death and binding to cetuximab in the absence of treatment. a Proliferation quantification in untreated SW48 WT, Mock and ST6Gal1 OE clones (OE 1 and OE 3) by flow cytometry analysis of bromodeoxyuridine (BrdU) staining. b Quantification of cell death by Annexin V/Propidium Iodate apoptosis assay in untreated SW48 WT, Mock and ST6Gal1 OE cell clones. c Cetuximab binding to cell surface EGFR in untreated SW48 WT, Mock and ST6Gal1 OE cell clones. Comparisons between multiple groups were made using one-way ANOVA, with Mock as control group. (PDF 2.62 MB)
ESM 2
(XLSX 9.36 MB)
Rights and permissions
About this article
Cite this article
Rodrigues, J.G., Duarte, H.O., Gomes, C. et al. Terminal α2,6-sialylation of epidermal growth factor receptor modulates antibody therapy response of colorectal cancer cells. Cell Oncol. 44, 835–850 (2021). https://doi.org/10.1007/s13402-021-00606-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-021-00606-z