Abstract—
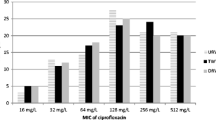
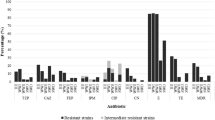
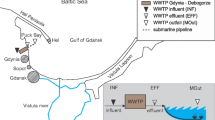
The work presents characterization of antibiotic-resistant strains isolated by direct plating of five samples collected at different treatment stages from the Pushchino water treatment facilities in April 2015. Primary analysis of resistance of the collection (~800 strains) to the following antibiotics was carried out: carbenicillin, kanamycin, streptomycin, amikacin, tobramycin, chloramphenicol, rifampicin, gentamicin, tetracycline, ceftazidime, cefepime, and meropenem. Antibiotic-resistant bacteria most common in the Pushchino wastewater treatment facilities were found to belong to the genera Pseudomonas and Alcaligenes. Occurrence of tetracycline resistance genes was investigated, and predominance of the tetA/tetC genes responsible for active transport of this antibiotic from the cell were found to be predominant among the studied strains. The strains containing the genes associated with type I integrons (intI1, qacE/qacEΔ1, and sul1) constituted 25% of the studied ones. Four Pseudomonas strains were found to contain the IncN plasmids, while seven strains of this genus contained plasmids of the P-9 incompatibility group (ε-subgroup). Three IncP-9 plasmids were conjugative and carried simultaneously the determinants of tetracycline, streptomycin, and gentamicin resistance, which has not been previously reported for the ε-subgroup of IncP-9 plasmids.




Similar content being viewed by others
REFERENCES
Ashbolt, N.J., Amézquita, A., Backhaus, T., Borriello, P., Brandt, K.K., Collignon, P., Coors, A., Finley, R., Gaze, W.H., Heberer, T., Lawrence, J.R., Larsson, D.G., McEwen, S.A., Ryan, J.J., Schönfeld, J., et al., Human Health Risk Assessment (HHRA) for environmental development and transfer of antibiotic resistance, Environ. Health Perspect., 2013, vol. 121, pp. 993‒1001.
Boronin, A.M., Diversity of Pseudomonas plasmids: to what extent?, FEMS Microbiol Lett., 1992, vol. 79, pp. 461‒467.
Carattoli, A., Plasmids and the spread of resistance, Int. J. Med. Microbiol., 2013, vol. 303, pp. 298–304.
Chaturvedi, P., Giri, B.S., Shukla, P., and Gupta, P., Recent advancement in remediation of synthetic organic antibiotics from environmental matrices: Challenges and Perspective, Bioresour. Technol., 2021, vol. 319, art. 124161.
Dalkmann, P., Broszat, M., Siebe, C., Willaschek, E., Sakinc, T., Huebner, J., Amelung, W., Grohmann, E., and Siemens, J., Accumulation of pharmaceuticals, Enterococcus, and resistance genes in soils irrigated with wastewater for zero to 100 years in central Mexico, PLoS One, 2012, vol. 7, art. e45397.
De Bruyne, K., Slabbinck, B., Waegeman, W., Vauterin, P., De Baets, B., and Vandamme, P., Bacterial species identification from MALDI-TOF mass spectra through data analysis and machine learning, Syst. Appl. Microbiol., 2011, vol. 34, pp. 20‒29.
Gillings, M.R., Gaze, W.H., Pruden, A., Smalla, K., Tiedje, J.M., and Zhu, Y.G., Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution, ISME J., 2015, vol. 9, pp. 1269‒1279.
Götz, A., Pukall, R., Smit, E., Tietze, E., Prager, R., Tschäpe, H., van Elsas, J.D., and Smalla, K., Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR, Appl. Environ. Microbiol., 1996, vol. 62, pp. 2621‒2628.
Greated, A. and Thomas, C.M., A pair of PCR primers for IncP-9 plasmids, Microbiology (Reading), 1999, vol. 145, pp. 3003‒3004. https://doi.org/10.1099/00221287-145-11-3003A
Hong, J.S., Yoon, E.-J., Song, W., Seo, Y.B., Shin, S., Park, M.-J., Jeong, S.H., and Lee, K., Molecular characterization of Pseudomonas putida group isolates carrying bla VIM-2 disseminated in a university hospital in Korea, Microb. Drug Resist., 2018, vol. 24, pp. 627‒634.
Izmalkova, T.Yu., Sazonova, O.I., Sokolov, S.L., Kosheleva, I.A., and Boronin, A.M., The P-7 incompatibility group plasmids responsible for biodegradation of naphthalene and salicylate in fluorescent pseudomonads, Microbiology (Moscow), 2005, vol. 74, pp. 290‒295.
Kraft, C.A., Timbury, M.C., and Platt, D.J., Distribution and genetic location of Tn7 in trimethoprim-resistant Escherichia coli, J. Med. Microbiol., 1986, vol. 22, pp. 125–131.
Kerrn, M.B., Klemmensen, T., Frimodt-Möller, N., and Espersen, F., Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance, J. Antimicrob. Chemother., 2002, vol. 50, pp. 513–516.
Larsson, D.G.J., de Pedro, C., and Paxeus, N., Effluent from drug manufactures contains extremely high levels of pharmaceuticals, J. Hazard. Mater., 2007, vol. 148, pp. 751–755.
Li, J., Zhang, H., Chen, Y., Luo, Y., and Zhang, H., Sources identification of antibiotic pollution combining land use information and multivariate statistics, Environ. Monit. Assess., 2016, vol. 188, p. 430.
Nguyen, F., Starosta, A.L., Arenz, S., Sohmen, D., Dönhöfer, A., and Wilson, D.N., Tetracycline antibiotics and resistance mechanisms, Biol. Chem., 2014, vol. 395, pp. 559–575.
Partridge, S.R., Kwong, S.M., Firth, N., and Jensen, S.O., Mobile genetic elements associated with antimicrobial resistance, Clin. Microbiol. Rev., 2018, vol. 31, pp. 1‒61.
Puckowsk, A., Mioduszewska, K., Łukaszewicz, P., Borecka, M., Caban, M., Maszkowska, J., and Stepnowsk, P., Bioaccumulation and analytics of pharmaceutical residues in the environment: a review, J. Pharm. Biomed. Anal., 2016, vol. 127, pp. 232–255.
Roberts, M.C., Mechanism of resistance for characterized tet and otr genes, http://faculty.washington.edu/marilynr/tetweb1.pdf (modified: Feb 2020).
Rowe-Magnus, D.A. and Mazel, D., The role of integrons in antibiotic resistance gene capture, Int. J. Med. Microbiol., 2002, vol. 292, pp. 115‒125.
Sambrook, J., Fritsch, E.F., and Maniatis, T., Molecular cloning: A Laboratory Manual, Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press, 1989, 2nd ed.
Sandvang, D., Aarestrup, F.M., and Jensen, L.B., Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104, FEMS Microbiol. Lett., 1998, vol. 160, pp. 37‒41.
Schlüter, A., Szczepanowski, R., Pühler, A., and Top, E.M., Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool, FEMS Microbiol. Rev., 2007, vol. 31, pp. 449–477.
Sevastsyanovich, Y.R., Krasowiak, R., Bingle, L.E.H., Haines, A.S., Sokolov, S.L., Kosheleva, I.A., Leuchuk, A.A., Titok, M.A., Smalla, K., and Tho-mas C.M., Diversity of IncP-9 plasmids of Pseudomonas, Microbiology (Reading), 2008, vol. 154, pp. 2929‒2941.
Svobodová, K., Semerád, J., Petráčková, D., and Novotný, Č., Antibiotic resistance in czech urban wastewater treatment plants: microbial and molecular genetic characterization, Microb. Drug Resist., 2018, vol. 24, pp. 830–838.
Wen, Q., Yang, L., Duan, R., and Chen, Z., Monitoring and evaluation of antibiotic resistance genes in four municipal wastewater treatment plants in Harbin, Northeast China, Environ. Pollut., 2016, vol. 212, pp. 34‒40.
Yu, Z., Michel, F.C., Jr., Hansen, G., Wittum, T., and Morrison, M., Development and application of real-time PCR assays for quantification of genes encoding tetracycline resistance, Appl. Environ. Microbiol., 2005, vol. 71, pp. 6926‒6933.
Yu, J., Liu, D., and Li, K., Influence of tetracycline on tetracycline-resistant heterotrophs and tet genes in activated sludge process, Curr. Microbiol., 2015, vol. 70, pp. 415–422.
Zhang, B., Yu, Q., Yan, G., Zhu, H., Xu, X.Y., and Zhu, L., Seasonal bacterial community succession in four typical wastewater treatment plants: correlations between core microbes and process performance, Sci. Rep., 2018, vol. 8, art. 4566. https://doi.org/10.1038/s41598-018-22683-1
Zhang, X.X. and Zhang, T., Occurrence, abundance, and diversity of tetracycline resistance genes in 15 sewage treatment plants across China and other global locations, Environ. Sci. Technol., 2011, vol. 45, pp. 2598–2604
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
Statement on the welfare of animals. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by D. Timchenko
Rights and permissions
About this article
Cite this article
Kosheleva, I.A., Izmalkova, T.Y., Sazonova, O.I. et al. Antibiotic-Resistant Microorganisms and Multiple Drug Resistance Determinants in Pseudomonas Bacteria from the Pushchino Wastewater Treatment Facilities. Microbiology 90, 187–197 (2021). https://doi.org/10.1134/S0026261721020077
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261721020077