Abstract
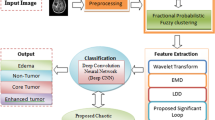
Many a times, the quality of Magnetic Resonance Imaging (MRI) based images get affected by the machines that are utilized to compute them. Hence, in several cases, it is necessary to process each image in order to obtain a corresponding better image so that the patient can be treated with ease and accuracy. According to traditional methods, this objective can be achieved by contrast enhancement or histogram stretching, etc. This paper give a naive approach that can enhance the MRI image, classify the brain tumor type and segment the tumor region for better analysis. For the enhancement, the concept of Discrete Wavelet is used in which an image is decomposed in four sub-bands and each sub-band is processed separately after which Inverse Discrete Wavelet Transform (IDWT) is applied in order to acquire a better quality image. Convolutional Neural Network (CNN) is used to classify the type of tumor present in MRI image. Lastly, after the tumor is detected, it is segmented by using a very efficient method known as K-means Clustering.















Similar content being viewed by others
REFERENCES
Bjoern H. Menze, Andras Jakab, Stefan Bauer, Jayashree Kalpathy-Cramer, Keyvan Farahani, Justin Kirby, Yuliya Burren, Nicole Porz, Johannes Slotboom, Roland Wiest, Levente Lanczi, Elizabeth Gerstner, Marc-André Weber, Tal Arbel, Brian B Avants, et al., “The multimodal brain tumor image segmentation benchmark (BRATS),” IEEE Trans. Med. Imaging 34 (10), 1993–2024 (2015).
M. Kistler, S. Bonaretti, M. Pfahrer, R. Niklaus, and Philippe Büchler, “The Virtual Skeleton Database: An open access repository for biomedical research and collaboration,” J. Med. Internet Res. 15 (11) (2013).
V. Kiruthika and M. M. Ramya, “Automatic segmentation of ovarian follicile using k-means clustering,” in Fifth International Conference on Signal and Image Processing (2014).
F. Kallel and A. B. Hamida, “Adaptive gamma correction based algorithm using DWT-SVD for non-contrast CT image enhancement,” IEEE Trans. NanoBiosci. 16 (8), 666–675 (2017). https://doi.org/10.1109/TNB.2017.2271350
X. Y. Wang, H. Y. Yang, and Z. K. Fu, “A new wavelet-based image denoising using undecimated discrete wavelet transform and least squares support vector machine,” Expert Syst. Appl. 37, 7040–7049 (2010).
K. Tsai, J. Ma, D. Ye, and J. Wu, “Curvelet processing of MRI for local image enhancement,” Int. J. Numer. Methods Biomed. Eng. 28 (6–7), 661–677 (2012).
T. Berger and J. O. Strömberg, “Exact reconstruction algorithms for the discrete wavelet transform using spline wavelets,” Appl. Comput. Harmonic Anal. 2 (4), 392–397 (1995).
N. Senthikumaran and J. Thimmiaraja, “Histogram equalization for image enhancement using MRI brain images,” in World Congress on Computing and Communication Technologies (2014).
Dipankar Ray, D. Dutta Majumder, and Amit Das, “Noise reduction and image enhancement of MRI using adaptive multiscale data condensation,” in 2012 1st International Conference on Recent Advances in Information Technology (RAIT) (IEEE, 2012).
Laurence C. Smith, Donald L. Turcotte, and Bryan L. Isacks, “Stream flow characterization and feature detection using a discrete wavelet transform,” Hydrol. Process. 12, 233–249 (1998).
F. Hsuan Cheng and Y. L. Chen, “Real time multiple object tracking and identification based on discrete wavelet transform,” Pattern Recognit. 39 (6), 1126–1139 (2006).
K. Dimililer and A. Ilhan, “Effect of image enhancement on MRI brain images with neural network,” in 12th International Conference on Application of Fuzzy Systems and Soft Computing; Procedia Comput. Sci. 102, 39–54 (2016).
S. Mukherjee, A. Adhikari, and M. Roy, “Malignant melanoma classification using cross-platform dataset with deep learning CNN architecture,” in Advances in Intellegent Systems and Computing (Springer, 2019), Vol. 922.
Z. Cao, J. C. Principe, Bing Ouyang, Fraser Dalgleish, and Anni Vuorenkoski, “Marine animal classification using combined CNN and hand-designed image features,” in OCEANS 2015 – MTS/IEEE Washington (IEEE, 2015).
J. Sun, C. Wan, F. Yu, and J. Liu, “Retinal image quality classification using fine-tuned CNN,” in Lecture Notes in Computer Science (Springer, 2017), Vol. 10554, pp. 126–133.
Saba S. Edris, Mohamed Zarka, Wael Ouarda, and Adel M. Alimi, “A fuzzy ontology driven context classification system using large-scale image recognition based on deep CNN,” in Sudan Conference on Computer Science and Information Technology (IEEE, 2017).
Zhiping Zhou, Xiaoxiao Zhao, and Shuwei Zhu, “K‑harmonic means clustering algorithm usinf feature weighting for color image segmentation,” Multimedia Tools Appl. 77, 15139–15160 (2018).
Jing-Hao Xue, W. Philips, A. Pizurica, and I. Lemahieu, “A novel method for adaptive enhancement and unsupervised segmentation of MRI brain image,” in 2001 IEEE International Conference on Acoustics, Speech, and Signal Processing. Proceedings (2001).
Van Huy Pham and Byung Ryong Lee, “An image segmentation approach for fruit defect detection using k-means clustering and graph-based algorithm,” Vietnam J. Comput. Sci. 2, 25–33 (2015).
Steven S. S. Poon, Rabab K. Ward, and Branko Palcic, “Automated image detection and segmentation in blood smears,” Cytometry 13, 766–774 (1992).
Jean Marie Vianney Kinani, Alberto J. Rosales-Silva, Francisco J. Gallegos-Funes, and Alfonso Arellano, “Fuzzy C-means applied to MRI images for an automatic lesion detection using image enhancement and constrained clustering,” in 2014 4th International Conference on Image Processing Theory, Tools and Applications (IPTA) (2014).
Nameirakpam Dhanachandra, Khumanthem Manglem, and Yambem Jina Chanu, “Image segmentation using k-means clustering algorithm and subtractive clustering algorithm,” in Eleventh International Conference on Information Processing (2015), Vol. 54, pp. 764–771.
G. Raghotham Reddy, K. Ramudu, Syed Zaheeruddin, and R. Rameshwar Rao, “Image segmentation using kernal fuzzy c-means clustering on level set method on noisy images,” in 2011 International Conference on Communications and Signal Processing (2011).
B. K. Tripathy, Avik Basu, and Sahil Govel, “Image segmentation using spatial intuitionistic fuzzy C means clustering,” in Image Segmentation Using Spatial İntuitionistic Fuzzy C Means Clustering (2014).
Yosep Aditya Wicaksono, Adhy Rizaldy, Sirli Fahriah, and Moch Arief Soeleman, “Improve image segmentation based on closed form matting using k-means clustering,” in 2017 International Seminar on Application for Technology of Information and Communication (iSemantic) (2017).
Anil D. Kumbhar and A. V. Kulkarni, “Magnetic resonant image segmentation using trained k-means clustering,” in 2011 World Congress on Information and Communication Technologies (2011).
J. C. Noordam and W. H. A. M. van den Broek, “Multivariate image segmentation based on geometrical guided fuzzy C-means clustering,” J. Chemom. 16 (1), 1–11 (2002).
H. P. Ng, S. H. Ong, K. W. C. Foong, P. S. Goh, and W. L. Nowinski, “Medical image segmentation using K-means clustering and improved watershed algorithm,” in SSIAI '06: Proceedings of the 2006 IEEE Southwest Symposium on Image Analysis and Interpretation (2006), pp. 61–65.
A. H. Hassin and M. M. Khudhair, “New algorithm for colour image segmentation using hybrid k-means clustering,” in 2nd Conference on Computer and Information Technology (2012).
I. Isa, S. N. Sulaiman, M. F. Abdullah, N. Md Tahir, M. Mustapha, and N. K. A. Karim, “New image enhancement technique for WMH segmentation of MRI FLAIR image,” in 2016 IEEE Symposium on Computer Applications & Industrial Electronics (ISCAIE) (2016).
Lei Zheng, J. C. Liu, A. K. Chan, and W. Smith, “Object based image segmentation using DWT/RDWT multiresolution Markov random field,” in 1999 IEEE International Conference on Acoustics, Speech, and Signal Processing. Proceedings. ICASSP99 (1999).
C. Huang and Z. Li, “Robust image segmentation using local robust statistics and correntropy-based k‑means clustering,” Opt. Lasers Eng. 66, 187–203 (2015).
Sugandhi Vij, Sandeep Sharma, and Chetan Marwaha, “Performance evaluation of color image segmentation using k means clustering and watershed technique,” in 2013 Fourth International Conference on Computing, Communications and Networking Technologies (ICCCNT) (2013).
Ritu Agrawal, Manisha Sharma, and Bikesh Kumar Singh, “Segmentation of brain lesions in MRI and CT scan images: A hybrid approach using k-means clustering and image morphology,” J. Inst. Eng. (India): Ser. B 99, 173–180 (2018).
P. Zamperoni, “Smoothing with adaptive rank order filters for early processing in image segmentation,” Int. J. Adapt. Control Signal Process. 9, 355–368 (1995).
Anupama K. Ingale, Nagaraj V. Dharwadkar, and Pratik Kodulkar, “Universal steganalysis using DWT and entropy features,” in 2016 International Conference on Signal and Information Processing (IConSIP) (IEEE, 2016).
S. W. Smith, The Scientist and Engineer’s Guide to Digital Signal Processing, 2nd ed. (California Technical Publishing, 1999).
Ashish M. Chikhalikar and Nagaraj V. Dharwadkar, “Enhanced brain tumor detection using segmentation based on discrete wavelet transform,” Int. J. Res. Advent Technol. 7 (3), 1225–1231 (2019).
Dipalee Gupta and Siddhartha Choubey, “Discrete wavelet transform for image processing,” Int. J. Emerging Technol. Adv. Eng. 4 (4), 598–602 (2014).
Amlan Jyoti, Mihir Narayan Mohanty, and Mallick Pradeep Kumar, “Morphological based segmentation of brain image for tumor detection,” in 2014 International Conference on Electronics and Communication Systems (ICECS) (IEEE, 2014).
k-means Clustering Algorithm. https://sites.google.com/site/dataclusteringalgoriths/k-means-clustering-algorithm.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
STATEMENT OF COMPLIANCE
This article does not contain any studies involving human participants performed by any of the authors.
Additional information

Ashish M. Chikhalikar obtained his B.Tech in Computer Science and Engineering in 2020 from Rajarambapu Institute of Technology, Shivaji University. He has worked on Signal Processing, Image Processing and Machine Learning. He has also completed his training on Blockchain Technology in Center of Artificial Intelligence and Robotics (CAIR), a branch of Defence Research and Development Organization (DRDO), India.

Nagaraj V. Dharwadkar obtained his BE in Computer Science and Engineering in 2000 from Karnataka University Dharwad, his M.Tech in Computer Science and Engineering in 2006 from VTU, Belgaum and PhD in Computer Science and Engineering in 2014 from National Institute of Technology, Warangal. He is a Professor and the Head of the Computer Science and Engineering Department at the Rajarambapu Institute of Technology, affiliated to Shivaji University, Islampur. He has 19 years of teaching experience at professional institutes across India and published 66 papers in various international journals and conferences. His area of research interest is multimedia security, image processing, data mining and machine learning.
Rights and permissions
About this article
Cite this article
Chikhalikar, A.M., Dharwadkar, N.V. Model for Enhancement and Segmentation of Magnetic Resonance Images for Brain Tumor Classification. Pattern Recognit. Image Anal. 31, 49–59 (2021). https://doi.org/10.1134/S1054661821010065
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1054661821010065