Abstract
The Eragrostis pilosa complex (Poaceae) comprises five widely distributed and regionally invasive species—E. albensis, E. amurensis, E. imberbis, E. multicaulis, and E. pilosa, distinguished by tiny and variable morphological characters and with so far unknown phylogenetic relationships. Recently, some doubts have been raised about the status of an invasive glandular morphotype occurring in Central Europe assigned either to E. amurensis or to E. albensis. Here, we addressed this issue by analysing morphology, internal transcribed spacers of nuclear ribosomal DNA, and five inter-simple sequence repeat markers. The genetic evidence supported closer relationship of this glandular morphotype to eglandular E. albensis, widely established in Central Europe, than to glandular E. amurensis described from Asia. We propose to adopt a new taxonomic treatment that E. albensis includes both eglandular and glandular individuals, and to classify the glandular ones as E. albensis var. scholziana M. Nobis & A. Wróbel var. nova. Currently this new taxon is known from a dozen of localities in Central Europe and is invasive in the lower section of the Oder River valley, whereas Eragrostis albensis var. albensis has already spread widely across Europe in riparian phytocenoses and anthropogenic habitats. Since probably the first registered records in 1940s, it has been observed in European part of Russia, Belarus, Ukraine, Poland, Slovakia, Czech Republic, Germany, Austria, the Netherlands, and its further invasion is likely to proceed. We provided distribution maps concerning spread dynamics of E. albensis in Europe from 1947 to 2020. In total, the species has been observed on over 1300 localities so far, most of which were found after 2000.
Similar content being viewed by others
Introduction
Biological invasions are currently considered one of the major global concerns threatening biodiversity and triggering economic losses (Pejchar and Mooney 2009; Pyšek and Richardson 2010). To prevent or at least mitigate such undesirable effects, immediate and precise identification of spreading newcomers is necessary in order to take appropriate countermeasures. Many alien species, however, belong to taxonomically problematic groups which makes their detection challenging and, therefore, usually time-delayed (Verloove 2010; Pyšek et al. 2013). Such cryptic invaders could be easily misidentified due to their morphological similarity to other closely related taxa and may remain unnoticed in the field for a long time. Moreover, insufficient knowledge about distribution of these organisms as well as routes and time of their spread may lead to difficulties while classifying them either as native or introduced in a particular region (Saltonstall 2002; Pyšek et al. 2013; Morais and Reichard 2017).
Nowadays, however, development and application of molecular markers may help to avoid confusion between invaders and other closely related native taxa, particularly those of conservation interest, as well as with other non-native taxa or species of economic importance (e.g. Newmaster and Ragupathy 2009; Cseke and Talley 2012; Wong et al. 2018; Martinez et al. 2020). The molecular approach can also be successfully applied to unravelling the population genetic structure of some invasive taxa, determining their routes of spread and sites of origin, locating their traces in environmental DNA, examining of their interactions and impact on native organisms, as well as identifying the features driving invasion success (e.g. Briski et al. 2011; Meyer et al. 2016; Hardouin et al. 2018; Fagúndez and Lema 2019). All of these issues are particularly relevant to biodiversity management and essential for the effective allocation of efforts and funding (Pyšek et al. 2013).
Genetic-based tools are thus appropriate candidates for application to taxonomically problematic groups such as grasses (Poaceae), a large (ca 12,000 species; Watson and Dallwitz 1992) monocot family of hard-to-identify plants occurring on all continents. Many grasses have been purposely propagated worldwide due to their agricultural and ornamental properties, while others have expanded their ranges spontaneously (David and Baruch 2000; Maillet and Lopez-Garcia 2000; Canavan et al. 2019). As a consequence, grasses include many examples of problematic invasive species, such as representatives of the following genera: Arundo, Bromus, Echinochloa, Eragrostis, Imperata, Pennisetum, Phalaris, Phragmites, Spartina (CABI 2020; Global Invasive Species Database 2020).
One of the largest groups of grasses is Eragrostis Wolf (lovegrasses), which comprises ca 400 species (Clayton et al. 2006) and is globally distributed from tropical to temperate regions, being mainly associated with dry, sandy, or human-disturbed areas. Therefore, some Eragrostis taxa are widely used as crop plants and forage (e.g. E. tef (Zucc.) Trotter; Cheng et al. 2017) or for erosion control (e.g. E. curvula (Schrad.) Nees; Lee et al. 2013), and have been intentionally introduced into many regions worldwide, where they have spread within both natural and anthropogenic habitats (Muranaka and Washitani 2004; Guzik and Sudnik-Wójcikowska 2005; Michalewska and Nobis 2005; Pagitz 2012). As a result, in many countries, the representatives of Eragrostis are now classified as alien casual or naturalised species, and regionally regarded as invasive (e.g. Pyšek et al. 2009; Yoshioka et al. 2010; Tokarska-Guzik et al. 2012).
In Central Europe, occurrence of Eragrostis could be dated back at least to the first half of the XIX century when, probably for the first time, E. minor Host was found in Silesia in 1838 within current borders of Wrocław in Poland (Fiek 1881). Since that time, over a dozen annual alien Eragrostis taxa have been observed in Central Europe and some of them have already become naturalised in this area (Špryňar and Kubát 2004; Guzik and Sudnik-Wójcikowska 2005; Scholz and Ristow 2005; Király et al. 2011; Medvecká et al. 2012; Hohla 2013). On a regional scale, two taxa are considered invasive species—E. minor in the Czech Republic (Danihelka et al. 2012) and E. albensis H. Scholz in Poland (Tokarska-Guzik et al. 2012; Dajdok et al. 2018). Other species, such as E. multicaulis Steud. and E. pilosa (L.) P. Beauv., are also spreading in Central Europe but have not been classified yet as invasive in this region (Scholz and Ristow 2005; Hohla 2006).
Recently, the occurrence of another species, E. amurensis Prob., was recorded in Central Europe along the Oder River valley in Germany in 2003 (B herbarium; Scholz and Ristow 2005) and in Poland in 2005 (WRSL, KRA herbarium; Kącki and Szczęśniak 2009) as well as along the Inn River valley in Upper Austria in 2013 (LI herbarium; Hohla 2013). Eragrostis amurensis was described as a new species from the Amur Province, Russian Far East on the basis of presence of numerous glands on a whole plant, especially on leaf sheaths, unlike mostly eglandular E. pilosa (Probatova and Sokolovskaya 1981). Eragrostis amurensis (= E. voronensis H. Scholz) as well as E. albensis, E. imberbis (Franch.) Prob., E. multicaulis, and E. pilosa belong to taxonomically problematic and widely distributed E. pilosa complex (Seregin 2012a). The most common characters used in the identification of these species are small differences in the morphology of the panicle and spikelets as well as presence or absence of glands and hairs on cauline leaf sheaths (Probatova 1985; Špryňar and Kubát 2004; Seregin 2012a). After delimitation of E. amurensis as a new species (Probatova and Sokolovskaya 1981), revision of herbarium materials and observations of its spread in the field have demonstrated that it occurs in a vast area of temperate Russia, Mongolia, Kazakhstan (Seregin 2012a), Tajikistan (Nobis et al. 2015; Wróbel et al. 2017), Belarus, Ukraine (Seregin 2012a; Parfenov 2013), and in Central Europe (Scholz and Ristow 2005; Kącki and Szczęśniak 2009; Hohla 2013).
However, Pagitz (2012) and Hohla (2013) have recently expressed some doubts about taxonomic treatment of the glandular E. amurensis specimens observed in Austria. Pagitz (2012) inclined to the view that glandular specimens from the North Tyrolean population found in 2005 (IB herbarium) could rather belong to E. albensis even though this species at that time was accepted as eglandular (Scholz 1995; Scholz and Ristow 2005). Pagitz (2012) suggested that E. albensis could probably include both eglandular and glandular morphotypes and put forward such a hypothesis for further studies. Hohla (2013) finally followed the taxonomic approach of Seregin (2012a) and classified the glandular plants from the Inn River valley in Upper Austria as E. amurensis, however, he highlighted their probable close relationship with the glandular specimens from North Tyrol. During our preliminary research focused on morphology of E. albensis and its spread in Poland, we observed that glandular plants from the Oder River valley, so far the only other known glandular population from Central Europe and previously also assigned to E. amurensis (Scholz and Ristow 2005; Kącki and Szczęśniak 2009), are extremely similar to eglandular E. albensis occurring in this region. Thus, our findings were in line with the previous speculations of Pagitz (2012) and Hohla (2013).
Eragrostis albensis differs from E. pilosa mostly in having shorter spikelet pedicels, slightly longer lemmas and glumes, more prominent lemma veins, usually no hairs at the apex of upper leaf sheaths, more scabrid and stiff panicle branches, and no verticillate branches in the lowest panicle node (Sholz 1995; Špryňar and Kubát 2004; Nobis and Nobis 2009). Eragrostis albensis was proposed to be regarded as a Central European young endemic taxon by Scholz (1995), at that time with known localities only from the Elbe and the Oder River valleys. Later, the revision of herbarium materials demonstrated that the occurrence of E. albensis in Germany could be dated back to 1982 when it was collected in Berlin (Scholz and Ristow 2005), while along the Elbe River valley the species had been documented since 1991 (Scholz and Ristow 2005), and from the Oder River valley since 1992 (Scholz 1995). Soon after, many other localities of E. albensis were reported from Central and Eastern Europe owing to the revision of herbaria material and new field observations. Probably the oldest collected specimen of E. albensis in the region is dated back to 1947 when the species was discovered along the Vistula River valley within current borders of Warsaw, Poland (WA herbarium; Guzik and Sudnik-Wójcikowska 2005). In 1968, E. albensis was for the first time found in Slovakia along the Danube River in Bratislava (Portal 2002) and in the same year in the Czech Republic in Prague (PR herbarium; Špryňar and Kubát 2004). These records are over 10 years older than the first findings in Germany dated back to 1982 (Scholz and Ristow 2005). Due to such pattern of distribution, Špryňar and Kubát (2004) proposed that E. albensis could rather be an invader in Central Europe, probably originating in other, more eastern parts of Eurasia. Nevertheless, the exact location of the origin spot of E. albensis was not determined. In Eastern Europe, where less research has been devoted to E. albensis, the first registered records of the species could probably be dated back to 1975 when it was collected in Bryansk, European part of Russia (MW herbarium; Seregin 2012b); later the species was also confirmed in Ukraine in 1997 (herbarium in Kiev; Gubar 2004) and Belarus (Guzik and Sudnik-Wójcikowska 2005). So far, the species was also found in Austria (Hohla 2006; Pagitz 2012) and the Netherlands (L herbarium). Despite increasing evidence about history of E. albensis spread, the exact location of its native range remains undetermined and a question whether it is autochtonous to Central Europe has not been fully answered yet (Špryňar and Kubát 2004; Pagitz 2012; Hohla 2013). Despite such circumstances, currently the leading approach is to recognize E. albensis in Central Europe as the alien invader (Špryňar and Kubát 2004).
An ongoing discussion about taxonomic treatment of Central European glandular Eragrostis specimens, so far either assigned to E. amurensis, E. amurensis-like plants or to E. albensis, has raised a question about their phylogenetic relationship as well as the current distribution and morphological variability of both E. amurensis and E. albensis (Scholz and Ristow 2005; Pagitz 2012; Hohla 2013). Due to such uncertainties and the observed invasive character of this glandular Eragrostis morphotype in Central Europe, the accurate identification of these grasses is particularly relevant for better understanding differentiation and routes of spread of the taxa within the E. pilosa complex which is essential for biodiversity management. Despite the growing interest in studies of Eragrostis (e.g. Cannarozzi et al. 2014; Carballo et al. 2019; Somaratne et al. 2019), molecular evidence concerning variability and relationships between the taxa from the E. pilosa complex still remains insufficient. This study aims to clarify the status of the glandular Central European Eragrostis morphotype from the complex by means of molecular markers. Due to the observed morphological character of these glandular plants as well as their occurrence in the close vicinity of eglandular E. albensis, we hypothesise that they are more closely related to eglandular E. albensis than to glandular E. amurensis and, therefore, that E. albensis comprises both eglandular and glandular morphotypes. To our knowledge, this research constitutes the first molecular phylogenetic insight into the E. pilosa complex with implications for its taxonomy and provides the most recent summary concerning spread of E. albensis in Central Europe.
Materials and methods
In this study we focused on five taxa from the E. pilosa complex (E. albensis, E. amurensis, E. imberbis, E. multicaulis, E. pilosa as well as Central European glandular Eragrostis specimens so far either assigned to E. amurensis or E. albensis). We also added five other Eragrostis taxa occurring in Eurasia: E. cilianensis (All.) Vignolo ex Janch., E. minor, E. pectinacea (Michx.) Nees, E. suaveolens A. K. Becker ex Claus, and E. virescens J. Presl. Extensive herbarium material was reviewed in order to determine morphological patterns among the examined Eragrostis taxa and to identify the most informative characters for the taxa from the E. pilosa complex. In total, more than 1500 specimens of Eragrostis were examined (materials deposited in the herbaria KRA, KRAM, LE, LI, SZUB, WA, WRSL and the herbarium of the University of Opole; acronyms according to Thiers 2020). Representative specimens were chosen for the morphological and molecular part of the research. For taxonomic comparisons, specimens of E. amurensis were purposely selected from the species′ core distribution area in Asia (specimens from Russian Siberia and Tajikistan).
Morphology
All examined specimens selected for morphological analyses were listed in “Online Appendix 1”. Measurements of panicles, spikelets, upper glumes, lower glumes, lemmas, and caryopses were carried out together with analyses of presence of glands, hairs, and prickles on different parts of culms (Online Appendix 2). Correlation between quantitative variables was checked using the Pearson correlation in RStudio version 1.1.423 (RStudio Team 2016) with R ver. 3.6.1 (R Core Team 2019). Variables characterised by a strong correlation coefficient (more than 0.90 or less than − 0.90) were then individually assessed; one of each pair was excluded from numerical analysis, since applying both might have influenced the final clustering arrangement unfavourably. Finally, the 20 most informative quantitative and qualitative characters (Online Appendix 2) were taken into consideration. The distance matrix was calculated by means of Gower′s similarity index using the proxy package in R (Meyer and Buchta 2019). The UPGMA (unweighted pair group method with arithmetic mean) cluster analysis was performed in R using the stats package (R Core Team 2019) to detect morphological patterns among the examined plants.
Molecular analyses
DNA from dried leaf tissue of herbarium specimens was isolated using a Genomic Mini AX Plant Spin (A&A Biotechnology, Poland) kit in accordance with the manufacturer′s protocol. Where necessary, the isolated DNA was purified using a gDNA Clean kit (Syngen, Poland). The purity and concentration of extracted DNA was assessed using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA). Only pure DNA with an A260/280 (DNA: protein ratio) value larger than 1.8 was used for the downstream analyses.
For ITS analysis we chose 25 individuals (Online Appendix 3). ITS1-5.8 S-ITS2 as a whole was amplified using forward primer N18L18: 5′-AAGTCGTAACAAGGTTTC-3′ (Wen and Zimmer 1996) and reverse primer ITS4: 5′-TCCTCCGCTTATTGATATGC-3′ (White et al. 1990). Amplification reactions were performed in a total volume of 25 μl, containing 10 ng of genomic DNA, 1 × final concentration of PCR DreamTaq Green Buffer (Thermo Scientific, USA), 1 U of DreamTaq Green DNA Polymerase (Thermo Scientific, USA), 0.12 mmol of dNTPs (Thermo Scientific, USA), 0.2 pmol of each primer, and 1 μg of bovine serum albumin (BSA). The PCR mixtures were heated at 94 °C for 3 min prior to 25–30 cycles of PCR amplification in a Veriti thermal cycler (Applied Biosystems, USA); one PCR cycle consisted of denaturation at 94 °C for 1 min, annealing of primers at 50 °C for 2 min, and extension at 72 °C for 2 min; following the last cycle, the PCR mixtures were incubated at 72 °C for 7 min. Reactions without DNA were used as negative controls. PCR products were sent to an external company (Genomed, Poland) for paired-end Sanger sequencing. The resulting sequences were manually verified and aligned using BioEdit ver. 7.0.5.3 (Hall 1999). One sequence of Eragrostis pectinacea was taken from GenBank (accession number: GU359301.1) and added to the dataset. One sequence of Enneapogon desvauxii (GenBank accession number: GU359339.1) was used as an outgroup.
As no studies had been published on the use of chloroplast markers for the E. pilosa complex, here we tested four cpDNA regions widely used in phylogenetic studies of angiosperms. The trnK-matK intron was amplified using primers trnK-3914F: 5′-TGG GTT GCT AAC TCA ATG G-3′ (Johnson and Soltis 1994) and matK-AR: 5′-CTG TTG ATA CAT TCG A-3′ (Osaloo et al. 1999). The trnCGCA-rpoB intergenic spacer was amplified using primers trnCGCAR: 5′-CAC CCR GAT TYG AAC TGG GG-3′ and rpoB: 5′-CKA CAA AAY CCY TCR AAT TG-3′, modified by Shaw et al. (2005) after Ohsako and Ohnishi (2000). The petL-psbE intergenic spacer was amplified using primers petL: 5′-AGT AGA AAA CCG AAA TAA CTA GTT A-3′ and psbE: 5′-TAT CGA ATA CTG GTA ATA ATA TCA GC-3′ (Shaw et al. 2007). The rpl32-trnLUAG intergenic spacer was amplified using primers trnLUAG: 5′-CTG CTT CCT AAG AGC AGC GT-3′ and rpL32-F: 5′-CAG TTC CAA AAA AAC GTA CTT C-3′ (Shaw et al. 2007). As a preliminary screening, selected specimens, representing E. albensis, E. amurensis, E. multicaulis, E. pilosa, and the glandular Eragrostis morphotype from Poland, were used to check the usefulness of selected cpDNA for genetic delimitation of species from the E. pilosa complex. All four cpDNA loci were amplified in a total volume of 25 μl, containing 10 ng of genomic DNA, 1 × final concentration of PCR DreamTaq Green Buffer (Thermo Scientific, USA), 1 U of DreamTaq Green DNA Polymerase (Thermo Scientific, USA), 0.12 mmol of dNTPs (Thermo Scientific, USA), 0.08 pmol of each primer, and 2 μg of BSA. The PCR mixtures were then heated at 80 °C for 5 min prior to 35 cycles of PCR amplification in a Mastercycler DNA thermal cycler (Eppendorf, Germany); one PCR cycle consisted of denaturation at 94 °C for 1 min, annealing of primers at 46 °C for 1 min, and extension at 72 °C for 2 min; following the last cycle, the PCR mixtures were incubated at 72 °C for 5 min. Reactions without DNA were used as negative controls. PCR products were sent to an external company (Genomed, Poland) for paired-end Sanger sequencing.
For analyses of ISSR (inter-simple sequence repeat; Ziętkiewicz et al. 1994) markers, 14 samples of Eragrostis were chosen (Online Appendix 3, Table 4). During a preliminary stage, the usefulness of 12 selected primers, including six previously designed for E. tef (Assefa et al. 2003), was assessed. Finally, five reproducible and scorable primers with polymorphic bands were chosen and used in PCR reactions in optimised annealing conditions (Table 1).
The PCR amplification of DNA fragments using the five ISSR primers was carried out in a total volume of 15 μl, containing 10 ng of genomic DNA, 1 × final concentration of PCR DreamTaq Green Buffer (for primers 811, 888, 889; Thermo Scientific, USA) or Taq Buffer with (NH4)2SO4 (for primers ubc836 and M2; Thermo Scientific, USA), 0.375 U (for 811, 888, 889) or 1 U (for ubc836) or 0.75 U (for M2) of DreamTaq Green DNA Polymerase (Thermo Scientific, USA), 3 pmol of dNTPs (Thermo Scientific, USA), 1.33 pmol of primer, 30 pmol of MgCl2 (Thermo Scientific, USA) for 811, 888, 889, and ubc836 or 45 pmol for M2.
The PCR mixtures were heated at 95 °C for 3 min prior to 33 cycles of PCR amplification in a DNA thermal cycler T100 (Bio-Rad, USA); one PCR cycle consisted of denaturation at 95 °C for 30 s, annealing of a primer at 53–58 °C (Table 1) for 30 s, and extension at 72 °C for 1 min; following the last cycle, the PCR mixtures were incubated at 72 °C for 10 min. For each primer, the PCR reaction was repeated to check reproducibility.
ISSR fragments were separated via electrophoresis for one hour at 100 V in 2% agarose gel with Midori Green DNA stain (Nippon Genetics, Germany) for visualisation. 1X TBE was used as a buffer solution. Subsequent imaging was carried out under UV light to confirm the amplification. The size of amplified fragments was estimated via comparison to a GeneRuler 100 bp Plus DNA Ladder (Thermo Scientific, USA) and scored in a binary format as either (1) present or (0) absent.
Molecular data analyses
Maximum-likelihood analysis (ML) of ITS marker was performed in PhyML 3.0 (Guindon et al. 2010). The best substitution model for the dataset was determined using SMS (Smart Model Selection; Lefort et al. 2017) with AIC (Akaike Information Criterion). The model GTR + I (general time-reversible model with a proportion of invariable sites) was indicated as most appropriate. Support values for the tree nodes were calculated using the approximate likelihood ratio test (aLRT; Anisimova and Gascuel 2006).
Bayesian Inference (BI) analysis was performed in MrBayes 3 (Ronquist and Huelsenbeck 2003). The most appropriate substitution model was selected using MrModeltest 2.3 (Nylander 2005) and PAUP* 4.0a166 (Swofford 2002). Model SYM + G (a symmetrical model with gamma distributed rate variation among sites) was indicated as the best and was set as lset nst = 6 rates = gamma, prset statefreqpr = fixed(equal). An MCMC simulation was set as a default for 1,000,000 generations, sampling one of every 500 generations, which sufficed to obtain the average standard deviation of split frequencies below 0.01 and the potential scale reduction factor of all parameters close to 1.0. Tracer 1.6.0 (Rambaut et al. 2014) was used to assess convergence and to ascertain whether all statistics were characterised by effective sample sizes (ESS) greater than 200. The first 25% of the iterations were discarded as a ′burn-in′ fraction; the remainder was used to construct the Bayesian consensus tree.
The trees were edited in TreeGraph 2 (Stöver and Müller 2010) and in MEGA ver. 7.0.26 (Kumar et al. 2016). Bayesian posterior probabilities (BPPs) ranged from 0.90 to 1 and support values from ML (MLs) higher than 0.7 (Hillis and Bull 1993) were regarded as sufficiently strong support for clades. Tree nodes with weak support, with both BPPs less than 0.9 and MLs less than 0.7, were collapsed and presented as unresolved (polytomy). To increase the clarity of the phylogram, the location of support values for each node was changed from the default tree output and adjusted manually using Inkscape ver. 3.
A binary ISSR matrix containing only polymorphic bands was analysed in RStudio version 1.1.423 (RStudio Team 2016) with R ver. 3.5.3 (R Core Team 2019). The distance matrix was calculated by means of the Jaccard similarity index using the proxy package (Meyer and Buchta 2019). A neighbour-joining tree and bootstrap analysis were performed using the ape package (Paradis and Schliep 2018). Bootstrap values higher than 0.7 were regarded as sufficiently strong support for clades (Hillis and Bull 1993). The tree was rooted by Eragrostis taxa from outside the E. pilosa complex.
Spread dynamics of Eragrostis albensis
Distribution maps of E. albensis spread in Europe were based on the specimens of this taxon preserved in the herbaria KRA, KRAM, LI, POZ, SZUB, WA, WRSL, information in online databases of herbarium specimens of B, BRNU, GJO, LZ, PRC, W, WU (JACQ—Virtual Herbaria; https://www.jacq.org/; accessed 2020-09-10), and MW (https://plant.depo.msu.ru/module/itemsearchpublic; accessed 2020-09-10), published localities (Online Appendix 5), ATPOL database (Zając A., Institute of Botany, Jagiellonian University; accessed 2020-09-14), GBIF database (https://doi.org/10.15468/dl.qc8cf9; accessed 2020-09-04), PLADIAS database (https://pladias.cz/en/; accessed 2020-09-10), observations of Wrzesień M. (Maria Curie-Skłodowska University in Lublin), and unpublished data from our field studies. Original GPS coordinates from a locality were used if possible. If GPS data was not available, geographic coordinates were approximated based on a description of a locality or a number of a grid unit (ATPOL, PLADIAS). Some regions including south-western Czech Republic, south-western Germany, central Poland, Slovakia, Hungary and Eastern Europe could be to some extent underestimated due to scarce or none data available. The list of all E. albensis localities which were included in the distribution maps is available in “Online Appendix 6”. Spread dynamics of E. albensis was divided into three time intervals and set arbitrarily to 1979, 1999, and 2020. The distribution maps were prepared in ArcGIS Desktop 10.8 with the ESRI basemap World Imagery. https://www.arcgis.com/home/item.html?id=10df2279f9684e4a9f6a7f08febac2a9.
Results
Morphology: key characters
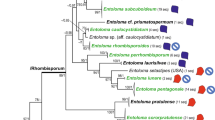
Analyses revealed five main distinct morphological groups within the E. pilosa complex (Fig. 1; Table 2). Two taxa, E. multicaulis and E. pilosa, were aggregated. Each formed its own cluster, which was clearly separated from the other taxa (Fig. 1, cluster IV and V, respectively). Eragrostis amurensis was linked in one cluster with Central European glandular specimens from the Inn River valley (Fig. 1, cluster I). Central European glandular specimens from the Oder River valley (Fig. 1, cluster III) were resolved as a sister cluster to an aggregation consisting of E. albensis and E. imberbis (Fig. 1, cluster II).
Dendrogram of the cluster analysis (UPGMA) of the Eragrostis pilosa complex based on 20 selected morphological characters (Online Appendix 2) using Gower′s similarity index. I—E. amurensis (green), glandular Eragrostis morphotype from the Inn River valley, Austria (red); II—E. albensis (dark blue), E. imberbis (light blue); III—glandular Eragrostis morphotype from the Oder River valley, Poland (pink); IV—E. multicaulis (orange); V—E. pilosa (yellow). A list of examined specimens can be found in “Online Appendix 1”
The most useful diagnostic characters of E. pilosa are: the presence of several verticillate branches in the lowest panicle node (in small individuals sometimes non-verticillate), presence of tufts of long hairs at the apex of all leaf sheaths, hairs in the panicle axils, and usually flexuous panicle branches. Eragrostis amurensis is very similar to E. pilosa, however, it has glands on leaf sheaths and blades. Eragrostis albensis has more robust and stiff panicle branches than E. pilosa, no verticillate branches in the lowest panicle node (if there are several branches, then they are grouped on one side of a panicle or in clusters opposite to each other, never arranged as a whorl), tufts of long hairs at the apex of usually only lower leaf sheaths, and hairs in panicle axils. Eragrostis imberbis is similar to E. albensis, however, it has longer pedicels of the lateral spikelets [E. imberbis: 2–5 mm long; E. albensis: (0.5–)1.0–2.5(–3.5) mm] and has usually several verticillate branches at the lowest panicle node. Eragrostis multicaulis has no hairs in panicle axils, no verticillate branches in the lowest panicle node, usually small robust panicle with branches diverging even to 90° at maturity, and no tufts of long hairs at the apex of all leaf sheaths (rarely, at most single long hair on some leaf sheaths).
The analyses showed that, when morphology was considered exclusively, Central European glandular specimens did not represent uniform patterns. Specimens from the Oder River valley referred morphologically to eglandular E. albensis to a greater extent than to glandular E. amurensis (Fig. 1). These plants had the longest lower glumes within the entire studied complex. Moreover, similarly as in E. albensis and E. imberbis (Fig. 1; Table 2), they had no tufts of long hairs at the apex of the uppermost leaf sheaths. Specimens from the Inn River valley reflected glandular E. amurensis to a greater extent than eglandular E. albensis (Fig. 1). These plants had less prominent glumes than specimens from the Oder River valley. In addition, they were characterised in part by long hairs at the apex of the uppermost leaf sheaths (Fig. 1; Table 2). All of the other morphological characters of Central European glandular morphotype overlapped considerably.
ITS region
The ITS sequences numbered 606 bp in all taxa from the E. pilosa complex and from 600 to 606 bp in other examined Eragrostis taxa. The tree topologies from the Bayesian inference method and the maximum-likelihood analysis based on ITS region were consistent. The Central European glandular Eragrostis morphotype was grouped in a clade with eglandular E. albensis, E. imberbis, and E. multicaulis (Fig. 2). The glandular specimens from the Oder River valley were identical with E. albensis. They differed, however, from the glandular specimen from the Inn River valley in Upper Austria by two substitutions (Table 3). All representatives of this clade, both glandular and eglandular differed from glandular E. amurensis by at least two substitutions (Table 3). In total, six sites were polymorphic in E. amurensis, E. albensis, E. imberbis, E. multicaulis, and the glandular morphotype from Central Europe. Eragrostis virescens appeared to be a sister clade to E. pilosa and other taxa from the E. pilosa complex. Eragrostis minor, E. suaveolens, E. pectinacea and E. cilianensis subsp. starosselskyi were resolved as more distantly related to the E. pilosa complex and formed a distinct clade (Fig. 2).
The phylogram from the Bayesian inference based on ITS1-5.8 S-ITS2 (nuclear ribosomal DNA). Numbers at nodes represent Bayesian posterior probabilities (upper) and ort values from maximum-likelihood analysis (lower). The scale bar represents substitutions per position. Abbreviations: Austria (AT), China (CN), Kyrgyzstan (KG), Mexico (MX), Poland (PL), Russia (RU), Tajikistan (TJ). *Central European glandular morphotype from the Eragrostis pilosa complex. All examined specimens are listed in “Online Appendix 3”
Chloroplast DNA regions
All studied chloroplast regions showed lower level of variation than ITS. Based only on the acquired sequences of good quality, several mutations were detected. In trnK-matK intron, one substitution was noted in E. amurensis (E2) in comparison to identical sequences of E. albensis (E6) and E. multicaulis (E17). In rpoB-trnCGCA intergenic spacer, two substitutions were observed in E. pilosa (E22) in comparison to E. albensis (E6). In petL-psbE intergenic spacer, one insertion was noted in E. amurensis (E2) in comparison to glandular morphotype from Central Europe (E34). In rpl32-trnL intergenic spacer, one deletion and nine substitutions were observed in E. pilosa (E22) in comparison to identical E. amurensis (E2), E. albensis (E6), E. multicaulis (E17), and glandular morphotype from Central Europe (E34).
ISSR markers
In total, the selected set of primers enabled the acquisition of 144 unique bands (characters). ISSR markers indicated that E. amurensis differed from the Central European glandular morphotype which was aggregated with eglandular E. albensis (Fig. 3; Table 4). Eragrostis imberbis appeared to be very closely related to E. albensis and the Central European glandular morphotype. Eragrostis amurensis, E. multicaulis, and E. pilosa were considered distinct lineages; however, relationships between these three taxa and E. albensis remained unresolved due to weak support (Fig. 3). The other three examined taxa, E. minor, E. virescens, and E. cilianensis subsp. starosselskyi, were regarded as distinct lineages outside the E. pilosa complex (Fig. 3).
Neighbour-joining tree of the ISSR profiles of examined Eragrostis taxa. The scale bar represents distance between specimens (calculated by means of the Jaccard similarity index). Numbers at nodes represent bootstrap values. Abbreviations: Austria (AT), China (CN), Kyrgyzstan (KG), Poland (PL), Russia (RU), Tajikistan (TJ). aCentral European glandular morphotype from the E. pilosa complex. All examined specimens are listed in “Online Appendix 3”
Taxonomic treatment
As a result of morphological and phylogenetic analyses of the Eragrostis pilosa complex, we propose to describe the glandular morphotype of Eragrostis albensis as follows:
Eragrostis albensis var. scholziana M. Nobis & A. Wróbel, var. nov. (Online Appendix 4).
TYPE: Western Poland, Lubuskie Province, Słubice, the Oder River valley, sandy banks on the right side of the river, 52°21′4.28″N/14°33′19.32″E, 17 m a.s.l., 26 August 2017, M. Nobis, A. Nowak, A. Wróbel s.n. (holotype KRA 474060!; isotypes KRA 474059, 474061–474064, 475309, 475310, 475312, 475313, 475315–475326, 475328, 475329, 475331, 475332, 475334, 475335, 475339–475346, 482318, 528738–528761!).
Diagnosis: Eragrostis albensis var. scholziana differs from eglandular E. albensis var. albensis in having glands on leaf sheaths keel and blade margins.
Other specimens studied (paratypes): western Poland, Lubuskie Province, Ługi Górzyckie, near the Oder River valley, field road, 52°32′2.27″N/14°37′20.33″E, 14 m a.s.l., 26 August 2017, M. Nobis, A. Nowak, A. Wróbel s.n. (KRA 473835, 482319!); western Poland, Lubuskie Province, Ługi Górzyckie, the Oder River valley, sandy banks on the right side of the river, 52º32′8.42″N/14º36′23.99″E, 10 m a.s.l., 26 August 2017, M. Nobis, A. Nowak, A. Wróbel s.n. (KRA 473831, 473836–473838, 47473–474075, 474078, 74079, 528718–528721!); north-western Poland, Zachodniopomorskie Province, between Czelin and Stary Błeszyn, the Oder River valley, sandy banks on the right side of the river, 52°44′41.56″N/14°21′32.96″E, 3 m a.s.l., 26 August 2017, M. Nobis, A. Nowak, A. Wróbel s.n. (KRA 473833, 473834, 474084, 475307, 475308, 482320!); north-western Poland, Zachodniopomorskie Province, Gozdowice, the Oder River valley, sandy banks on the right side of the river, 52°45′49.94″N/14°19′9.17″E, 5 m a.s.l., 26 August 2017, M. Nobis, A. Nowak, A. Wróbel s.n. (KRA 475302–475305, 528711–528717!); north-western Poland, Zachodniopomorskie Province, Osinów Dolny, the Oder River valley, sandy banks on the right side of the river, 52°51′26.94″N/14°8′19.61″E, 1 m a.s.l., 26 August 2017, M. Nobis, A. Nowak, A. Wróbel s.n. (KRA 473823, 474067–474070, 474119, 528722–528738!); Kunice, Odra, alluvia, 3 September 2005, Z. Kącki s.n. (WRSL!); Porzecze (1), Odra, 3 September 2005, Z. Kącki s.n. (WRSL!); Czelin, Odra, alluvia, 2 September 2005, Z. Kącki s.n. (WRSL!); Szumiłowo, Odra (3), 3 September 2006, Z. Kącki s.n. (WRSL!); Górzyca (1), Odra, alluvia, 3 September 2005, Z. Kącki s.n. (WRSL!); Austria, Inn River region, Hagenauer Bucht St. Peter am Hart [Österreich, Oberösterreich, Innviertel, St. Peter am Hart, Hagenauer Bucht, junge Anlandungen an der Südwestseite der “Kellerinsel”], 13°5′47″E/48°16′32″, MTB:7744/2, Unschärferadius: 100 m, 334 m, 15 September 2013, M. Hohla (LI 75143!); Austria, Inn River region, Hagenauer Bucht St. Peter am Hart [Österreich, Oberösterreich, Innviertel, St. Peter am Hart, Hagenauer Bucht, Insels SE des Leitdammdurchstiches], 13°4′36″E/48°16′14″N, MTB:7744/1, Unschärferadius: 100 m, 335 m, 15 September 2013, M. Hohla (LI 751048!).
Spread of Eragrostis albensis in Central Europe
Since the first confirmed record of E. albensis in Europe (1947, Warsaw, Poland), the species has been observed on over 1250 localities in Central Europe, over 50 in Eastern Europe, and over 10 in Western Europe (Fig. 4). From 1947 to 1979 only three localities were confirmed in Central Europe. In 1980–1999, a considerable spread of E. albensis was noted mainly along the Elbe River valley in Germany and along the Vistula River valley in Poland. The species was also found on several localities along the Oder River valley in Germany and on a few other localities in anthropogenic habitats. In the last 20 years, E. albensis propagated further especially along the Elbe River and the Oder River (Fig. 5) valleys. It was also observed in many new localities in anthropogenic habitats, particularly along roadsides in Upper Austria, North Tyrol, south and east Poland as well as along rail in south-eastern and east Poland (Fig. 4).
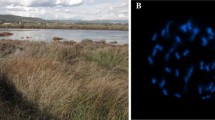
Riparian communities from the Bidentetea tripartiti class along the Oder River, invaded by Eragrostis albensis var. scholziana (white arrow) in Słubice, Poland, 52°21′4.28″ N, 14°33′19.32″ E (a); mature generative shoots of eglandular E. albensis var. albensis on sandy banks of the Oder River near Leśna Góra, Poland, 52°01′52.2″ N, 15°38′16.9″ E (b); photographs taken 26 August 2017 by A. Wróbel
Discussion
The place of origin, distribution and differentiation of the taxa from the E. pilosa complex has been already lively discussed but so far with no support of molecular evidence (Špryňar and Kubát 2004; Guzik and Sudnik-Wójcikowska 2005; Pagitz 2012; Seregin 2012a; Hohla 2013). This study indicates that glandular specimens of lovegrasses from Central Europe are more closely related to eglandular E. albensis than to glandular E. amurensis as was previously suggested (Scholz and Ristow 2005). Therefore, as opposite to the typical eglandular E. albensis var. albensis, we proposed to delineate a glandular variety of this species under the name E. albensis var. scholziana. Eglandular E. albensis var. albensis is much more widespread and occurs widely across Central, Eastern, and part of Western Europe, and is already invasive along several Central European rivers, mainly the Elbe, the Oder and the Vistula. On the other hand, the glandular E. albensis var. scholziana is so far known only from a dozen of localities in Central Europe and currently is invasive only in the lower section of the Oder River valley.
Results of our work provide another example that illustrates a wide problem concerning a time-delayed identification of spreading organisms which have broad distribution and complicated taxonomy. Such cryptic invaders may remain overlooked or misidentified for a long time, very often until they start to attract more attention after becoming established or problematic in a particular area (Saltonstall 2002; Gerlach et al. 2009; Wong et al. 2018). Another challenge is that lack of solid evidence about history of spread and a place of origin may lead to confusion while classifying a taxon as either native or alien in a specific region (Pyšek et al. 2013; Morais and Reichard 2017).
Unresolved native ranges of E. albensis and E. amurensis
Eragrostis albensis native range has not been conclusively resolved. Scholz (1995) proposed that E. albensis could be a Central European young endemic taxon originated as a result of a rapid evolutionary process from a single closely related Eragrostis individual introduced by accident from the eastern countries. However, the leading hypothesis suggests that it could rather be an invader, which originated outside Central Europe and invaded this region from the east borders (Špryňar and Kubát 2004).
Like for Eragrostis albensis, E. amurensis native range has not been resolved yet. Lomonosova (2000) presumed that its westernmost borders reach only Novosibirsk Province, Western Siberia (Russia). On the other hand, Seregin (2012a) proposed to determine native range of E. amurensis as reaching further to the west, as far as to SE Belarus and Central and Eastern Ukraine. Our study indicates that previous records of E. amurensis from Central Europe are invalid and, therefore, its known distribution does not reach further than to Eastern Europe. However, taking also into account that E. albensis is established in European part of Russia, Belarus and Ukraine (Sukhorukov 2011; Seregin 2012a,b), there is a need for a revision of glandular specimens from Eastern Europe and Western Siberia to determine if these specimens indeed represent E. amurensis or maybe rather E. albensis var. scholziana, or even both taxa. Such research could shed more light on actual distribution and native ranges of these two species in Eurasia.
Eragrostis albensis introduction route to Europe
Probably, the first localities of E. albensis in both Eastern and Central Europe were most likely overlooked. Therefore, the data from existing herbarium material could be biased to some extent and should be analysed cautiously. Since the first known registered records in 1947 from Warsaw in Poland, E. albensis has become widely established in Central Europe and so far it has been noted in hundreds of localities, most commonly in Germany and Poland. One probable scenario of invasion is that E. albensis could have probably invaded Central Europe from the east (Špryňar and Kubát 2004), firstly through anthropogenic habitats and then started to penetrate adjacent riparian communities most likely near roads and bridges (Michalewska and Nobis 2005). Taking into consideration the time of the first confirmed records in Central Europe, an introduction of the species to this region could probably be attributed to military actions during the Second World War in 1940s and accidental dispersal of E. albensis seeds from the territory of Russia towards Central European countries. After that, E. albensis could have spread further simultaneously through rail, road traffic and along riversides, which was enhanced by dispersal of seeds in sand and gravel gathered along rivers and used in building areas as well as for road maintenance during winter (Michalewska and Nobis 2005). Moreover, floods in XX century might have promoted its rapid propagation along rivers (Guzik and Sudnik-Wójcikowska 2005). In the last 20 years, the species has considerably spread especially in anthropogenic habitats, most likely due to constantly growing road traffic and use of sand from the river banks for road maintenance.
Eragrostis albensis invaded habitats and spreading in Central Europe
Eragrostis albensis is a therophyte which produces numerous small seeds, ca 0.8 × 0.4 mm, which could be dispersed by watercourse, human activities, animals or wind over long distances. Along rivers, E. albensis prefers open and sparse alluvial communities including sandy, gravelly and muddy areas on floodplain terraces or river banks exposed during summer low water levels (Dajdok et al. 2018). It occurs abundantly in alluvial habitats and forms dense extensive stands which considerably limit the area for other plants. As a result, E. albensis has already outcompeted some of the native species in the region mainly along the Oder (Fig. 4), Elbe, and Vistula River valleys where it has invaded riparian phytocoenoses, including those protected by NATURA 2000—Isoëto-Nanojuncetea, code 3130 and Bidentetea tripartiti, code 3270 (Guzik and Sudnik-Wójcikowska 2005; Krumbiegel 2008; Kącki and Szczęśniak 2009; Dajdok et al. 2018). Moreover, the species could regionally have a negative impact on other therophytes growing on river banks. Spreading E. albensis has already been identified as a potential threat to species such as Corrigiola litoralis L., Dichostylis micheliana (L.) Nees, Lindernia procumbens (Krock.) Philcox, and Lythrum hyssopifolia L. (Kącki and Szczęśniak 2009; Jackowiak et al. 2014; Dajdok et al. 2018).
According to our research, E. albensis var. scholziana occurs in much fewer localities compared to E. albensis var. albensis. Glandular variety occurs both along roadsides and rivers (Scholz and Ristow 2005; Kącki and Szczęśniak 2009; Pagitz 2012; Hohla 2013), and so far, it exhibits invasive potential along the Oder River valley, where it grows abundantly and usually with no admixtures of the typical eglandular morphotype. A spread of E. albensis var. albensis has been widely observed in Central Europe along rivers, mainly the Oder, Elbe, and Vistula River valleys as well as in anthropogenic habitats including roadsides, railway tracks, and pavements (Guzik and Sudnik-Wójcikowska 2005; Michalewska and Nobis 2005; Pagitz 2012; Wróbel and Nobis 2017). Taking into account the dynamics of E. albensis propagation, it seems very likely that its spread will continue and new localities will be reported soon. Moreover, similarly to other species spreading along motorways and main roads (e.g. Ambrosia artemisiifolia L., Cochlearia danica L., Dittrichia graveolens (L.) Greuter, Sagina maritima G.Don, Senecio inaequidens DC.; Brandes 2009; Zając and Zając 2019), it is probably a matter of time that E. albensis will be observed also in other countries which are adjacent to its currently known distribution.
Glandular morphotypes in the E. pilosa complex
The molecular evidence obtained in this study suggests that the presence of glands as well as hairs, in some cases, may not constitute a species-specific character in Eragrostis. Instead, it may rather be regarded as variability within one species. The species-specificity of the morphology, distribution, and abundance of glands as well as hairs has been already the subject of lively debate in botany and is far from resolved (e.g. Van den Borre and Watson 1994; Taia 2006; Ciccarelli et al. 2007; Pagitz 2012; Rola et al. 2019). In the genus Eragrostis, the presence of glands (crateriform-like structures, pits, or bands on various parts of a plant) was recognised as a useful and diagnostic morphological character enabling the identification of particular taxa, including in the field (Tutin 1980; Van den Borre and Watson 1994; Peterson 2003; Shouliang and Peterson 2006; Giraldo-Cañas et al. 2012). In the E. pilosa complex, the typical E. pilosa is considered completely eglandular or at least eglandular on leaf sheaths and blades. It may only sporadically have few glands on a panicle axis or below culm nodes (Koch 1974; Peterson 2003; Giraldo-Cañas et al. 2012; Seregin 2012a). In the complex, presence of glands on leaf sheaths has been related to three taxa so far – E. perplexa delimited in the USA as well as E. amurensis and E. voronensis described from Russia. Densely glandular plants observed in America were proposed as Eragrostis perplexa L.H.Harv., (Harvey 1954), subsequently, lowered to the rank of a variety by Koch (1974) as Eragrostis pilosa var. perplexa (L.H.Harv.) S.D.Koch and regarded by him as restricted to North America. This taxon was distinguished mainly on the basis of presence of abundant glands scattered on a whole plant and characterised by longer glumes, lemmas and caryopsis in comparison to the typical E. pilosa (Koch 1974; Peterson 2003). In Eurasia, two species with glandular leaf sheaths were described—E. amurensis (Probatova and Sokolovskaya 1981) and E. voronensis (Scholz 2010), however, the latter was considered a synonym of the former species after a taxonomic revision (Seregin 2012a). As a consequence, all individuals morphologically assigned to the E. pilosa complex in Eurasia but characterised by possession of glands on leaf sheaths have been identified as E. amurensis (Scholz and Ristow 2005; Kącki and Szczęśniak 2009; Seregin 2012a).
Nevertheless, our findings support the hypothesis of Pagitz (2012) that E. albensis could also express both eglandular and glandular morphotypes which implies that presence of glands on leaf sheaths should no longer be treated as an exclusive character of E. amurensis in the E. pilosa complex in Eurasia. Moreover, E. albensis may be variable across its range and could comprise more than one genetic lineage as the glandular specimens from the Inn River valley in Upper Austria had a slightly different genetic profile and morphology in comparison to plants from the Oder River valley. The next step should be to examine glandular specimens of Pagitz (2012) from North Tyrol, Austria using the ITS marker to determine if they represent the same genetic pattern as Upper Austrian population or form another evolutionary lineage of E. albensis.
Although we did not find any hints of recent hybridisation between taxa from the E. pilosa complex based on the ITS sequences, more data is needed to test this possibility. In addition, the potential crossing between taxa from the E. pilosa complex and more distant Eragrostis species should be verified. Probatova and Sokolovskaya (1981) speculated about putative origin of E. amurensis and attributed it to hybridisation between eglandular E. pilosa and glandular E. minor. However, this hypothesis was not supported by molecular evidence by Probatova and Sokolovskaya (1981) and would require further investigation.
Hairy plants in the E. pilosa complex
The characters related to hairs have been widely adopted as diagnostic in the E. pilosa complex. A presence of tufts of long hairs on lower leaf sheaths has been indicated as a diagnostic for E. albensis, whereas E. multicaulis has been generally accepted as not having tufts of long hairs and E. pilosa as having them on all leaf sheaths (Scholz 1995; Špryňar and Kubát 2004; Michalewska and Nobis 2005; Nobis and Nobis 2009; Seregin 2012a). During our revision, we found that all specimens of E. albensis from Poland, both glandular and eglandular, had tufts of long hairs at the apex only of lower leaf sheaths and did not have long hairs at the uppermost leaf sheaths. In addition, they all had hairs in panicle axils. We also observed that E. multicaulis examined from different countries did not have hairs in panicle axils and tufts of long hairs at the apex of leaf sheaths or at most only single long hairs on some leaf sheaths. Such pattern is therefore specific for many populations.
Pagitz (2012) also raised a problem related to the identification of dwarf specimens of E. albensis which could be confused with E. multicaulis which is regularly smaller and has shorter panicles than E. albensis. Such difficulty was also experienced by Scholz who suggested to identify small plants from the Oder River valley as E. multicaulis and bigger ones as E. albensis (specimens in WRSL herbarium) even though both morphotypes had tufts of long hairs at the apex of lower leaf sheaths. However, Pagitz (2012) noted that E. albensis in North Tyrol, Austria partly had tufts of long hairs at the uppermost leaf sheath (which makes these individuals more similar to E. pilosa than to E. albensis). Similarly, the glandular specimens from the Inn River valley in Upper Austria partly had tufts of long hairs at the uppermost leaf sheaths. That could suggest that there is variability in the expression of hairs in the species from the E. pilosa complex which could complicate their identification. As a consequence, collected evidence implies that presence of both hairs and glands could represents regionally variable patterns and, therefore, should rather be treated more cautiously in the taxonomic treatment of the E. pilosa complex.
A case of Eragrostis imberbis
The problem concerning classification and distribution of taxa from the E. pilosa complex becomes even more complicated if we consider one other taxon, E. imberbis. It was described from Russian Far East and was characterised by huge panicles usually longer than the rest of the culm, distinctly long spikelet pedicels, scabrid panicle branches, and eglandular leaf sheaths (Tzvelev 1976; Probatova 1985). Seregin (2012a,b) suggested that Asian E. imberbis distributed across Russian Far East and south Siberia might possibly be conspecific with European E. albensis and that these two taxa could be one species broadly distributed across Eurasia. Eragrostis albensis has never been reported from Asia, however, according to the latest revision, it appeared that it is not possible to indicate clear morphological differences between E. albensis and E. imberbis (Seregin 2012a,b). Eragrostis imberbis can be distinguished from E. pilosa mainly by having densely scabrous panicle branches and longer lemma (Probatova 1985; Seregin 2012a), and these two diagnostic differences are similar to those between E. albensis and E. pilosa. This study confirmed that specimens from Russian Far East, identified as E. imberbis, resembled morphology of European E. albensis to a large extent. Eragrostis imberbis differed only slightly from E. albensis in longer pedicels of lateral spikelets and by having usually several verticillate branches at the lowest panicle node (Table 2). Our molecular analyses resolved E. imberbis as a distinct although closely related lineage to E. albensis. The next step should be to use more sensitive, genome-wide approach and population sampling across Eurasia to get a better insight into the evolutionary history and dispersal dynamics at the population level of species from the E. pilosa complex.
Conclusions
Molecular analyses based on ITS and ISSR markers have indicated that the E. pilosa complex is genetically variable across Eurasia and there is a possibility to determine species- and lineage-specific genetic apomorphies. The first molecular insight to the complex indicated that E. albensis deviates from E. pilosa in many traits and, therefore, deserves to be treated as a separate taxon. The present study also revealed that glandular Central European morphotype, so far classified as E. amurensis or E. albensis, is genetically more similar to eglandular E. albensis, widely established in Central Europe, than to glandular E. amurensis described from Asia. Here, we propose to adopt a new taxonomic treatment that E. albensis includes both eglandular and glandular individuals, and classify the glandular variety as E. albensis var. scholziana. Our study implies that presence of glands on leaf sheaths should not be treated as an exclusive character of E. amurensis within the E. pilosa complex in Eurasia and, therefore, more attention should now be paid to glandular Eragrostis plants during identification.
Since the first confirmed records in 1947 till now, E. albensis has spread considerably in Central Europe along river valleys, roadsides and railway tracks. Its progressive invasion may potentially pose a threat to native riparian phytocenoses, therefore, regular biomonitoring would be recommended in order to control its impact on local alluvial communities and populations of rare species occurring there.
This study could be a starting point for further large-scale research which is needed to get a full picture of variability within the E. pilosa complex across Northern Hemisphere. It would be especially essential to determine whether the glandular specimens recorded from Eastern Europe and Western Siberia do not actually represent the glandular E. albensis var. scholziana. Further investigation could also address the question if E. albensis and E. imberbis, resolved as distinct lineages in this study, constitute two closely related but different species.
GenBank accessions
ITS1-5.8 S-ITS2 of nuclear ribosomal DNA.
Eragrostis albensis: MT344699 (E5), MT344698 (E6), MT344694 (E11); Eragrostis albensis var. scholziana: MT344702 (E3), MT344700 (E4), MT344691 (E14), MT344678 (E32); Eragrostis amurensis: MT344701 (E2), MT344697 (E7), MT344680 (E29); Eragrostis cilianensis subsp. starosselskyi: MT344696 (E9), MT344692 (E13); Eragrostis imberbis: MT344682 (E26), MT344681 (E28); Eragrostis minor: MT344695 (E10), MT344693 (E12); Eragrostis multicaulis: MT344688 (E17), MT344687 (E18), MT344679 (E31); Eragrostis suaveolens: MT344686 (E19); Eragrostis pilosa: MT344685 (E20), MT344684 (E22), MT344683 (E23); Eragrostis virescens: MT344690 (E15), MT344689 (E16).
Chloroplast DNA
petL-psbE intergenic spacer, partial sequence—Eragrostis albensis var. scholziana: MW019440 (E34); Eragrostis amurensis: MW019441 (E2).
rpL32 gene, partial cds; and rpl32-trnL(UAG) intergenic spacer, partial sequence—Eragrostis albensis: MW036251 (E6); Eragrostis albensis var. scholziana: MW036254 (E34); Eragrostis amurensis: MW036250 (E2); Eragrostis multicaulis: MW036252 (E17); Eragrostis pilosa MW036253 (E22).
trnC(GCA)-rpoB intergenic spacer, partial sequence; and (rpoB) gene, partial cds –
Eragrostis albensis: MW036255 (E6); Eragrostis pilosa MW036256 (E22).
tRNA-Lys (trnK) gene, intron, partial sequence; and matK gene, partial cds—Eragrostis albensis: MW036258 (E6); Eragrostis amurensis: MW036257 (E2); Eragrostis multicaulis: MW036259 (E17).
Data availability
Data are available in the electronic appendices, and are also available from the corresponding author on reasonable request.
References
Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55(4):539–552. https://doi.org/10.1080/10635150600755453
Assefa K, Merker A, Tefera H (2003) Inter simple sequence repeat (ISSR) analysis of genetic diversity in tef [Eragrostis tef (Zucc.) Trotter]. Hereditas 139:174–183. https://doi.org/10.1111/j.1601-5223.2003.01800.x
Brandes D (2009) Autobahnen als Wuchsorte und Ausbreitungswege von Ruderal- und Adventivpflanzen [Motorways as habitats and conduits for ruderal plants and alien species]. Braunschw Naturkdl Schr 8(2):373–394
Briski E, Cristescu ME, Bailey SA, MacIsaac HJ (2011) Use of DNA barcoding to detect invertebrate invasive species from diapausing eggs. Biol Invasions 13:1325–1340. https://doi.org/10.1007/s10530-010-9892-7
CABI (2020) Invasive species compendium. Wallingford, UK: CAB International. www.cabi.org/isc. Accessed 01 Jan 2020
Canavan S, Pyšek P, Packer JG et al (2019) Tall-statured grasses: a useful functional group for invasion science. Biol Invasions 21:37–58. https://doi.org/10.1007/s10530-018-1815-z
Cannarozzi G, Plaza-Wüthrich S, Esfeld K, Larti S, Wilson YS, Girma D, de Castro E, Chanyalew S, Blösch R, Farinelli L, Lyons E, Schneider M, Falquet L, Kuhlemeier C, Assefa K, Tadele Z (2014) Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef). BMC Genomics 15:581. https://doi.org/10.1186/1471-2164-15-581
Carballo J, Santos BACM, Zappacosta D, Garbus I, Selva JP, Gallo CA, Díaz A, Albertini E, Caccamo M, Echenique V (2019) A high-quality genome of Eragrostis curvula grass provides insights into Poaceae evolution and supports new strategies to enhance forage quality. Sci Rep 9:10250. https://doi.org/10.1038/s41598-019-46610-0
Cheng A, Mayes S, Dalle G, Demissew S, Massawe F (2017) Diversifying crops for food and nutrition security—a case of teff. Biol Rev 92:188–198. https://doi.org/10.1111/brv.12225
Ciccarelli D, Garbari F, Pagni AM (2007) Glandular hairs of the ovary: a helpful character for Asteroideae (Asteraceae) taxonomy? Ann Bot Fenn 44(1):1–7
Clayton WD, Vorontsova MS, Harman KT, Williamson H (2006) GrassBase—The Online World Grass Flora. http://www.kew.org/data/grasses-db.html. Accessed 11 Sep 2019
Cseke LJ, Talley SM (2012) A PCR-based genotyping method to distinguish between wild-type and ornamental varieties of Imperata cylindrica. J Vis Exp 60:e3265. https://doi.org/10.3791/3265
Dajdok Z, Nobis M, Sudnik-Wójcikowska B (2018) Eragrostis albensis. The analysis of the degree of invasiveness of alien species in Poland with an indication of species that significantly threaten native flora and fauna and a proposal of strategic actions in terms of the possibility of eradication. Analysis of the routes of unintentional introduction or spread of invasive alien species along with the development of action plans for priority roads. GDOŚ, Poland. http://projekty.gdos.gov.pl/files/artykuly/127056/Eragrostis-albensis_milka-polabska_KG_WWW_icon.pdf
Danihelka J, Chrtek J, Kaplan Z (2012) Checklist of vascular plants of the Czech Republic. Preslia 84:647–811
David GW, Baruch Z (2000) African grass invasion in the Americas: ecosystem consequences and the role of ecophysiology. Biol Invasions 2:123–140
Fagúndez J, Lema M (2019) A competition experiment of an invasive alien grass and two native species: are functionally similar species better competitors? Biol Invasions 21:3619–3631. https://doi.org/10.1007/s10530-019-02073-y(0123456789(),-volV)(01234567
Fiek E (1881) Flora von Schlesien preussischen und österreichischen Anteils, enthaltend die wildwachsenden, verwilderten und angebauten Phanerogamen und Gefäss-Cryptogamen. Verlag, Breslau, p 514. https://www.sbc.org.pl/dlibra/publication/11618/edition/10735
Gerlach JD, Bushman BS, McKay JK, Meimberg H (2009) Taxonomic confusion permits the unchecked invasion of vernal pools in California by low mannagrass (Glyceria declinata). Invasive Plant Sci Manag 2(1):92–97. https://doi.org/10.1614/IPSM-08-095.1
Giraldo-Cañas D, Peterson PM, Sánchez Vega I (2012) The genus Eragrostis (Poaceae: Chloridoideae) in northwestern South America (Colombia, Ecuador and Peru): morphological and taxonomic studies. Biblioteca José Jéronimo Triana 24:1–195
Global Invasive Species Database (2020) http://www.iucngisd.org/gisd/. Accessed 01 January 2020
Gubar LM (2004) New for the flora of Male Polissia species of vascular plants. Ukr Bot Zhurn 61(1):70–74
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321. https://doi.org/10.1093/sysbio/syq010
Guzik J, Sudnik-Wójcikowska B (2005) Critical review of species of the genus Eragrostis in Poland L Frey Eds Biology of grasses W Szafer, Institute of Botany Polish Academy of Sciences, Kraków, pp 45–58
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hardouin EA, Andreou D, Zhao Y, Chevret P, Fletcher DH, Britton JR, Gozlan RE (2018) Reconciling the biogeography of an invader through recent and historic genetic patterns: the case of topmouth gudgeon Pseudorasbora parva. Biol Invasions 20:2157–2171. https://doi.org/10.1007/s10530-018-1693-4
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192. https://doi.org/10.1093/sysbio/42.2.182
Hohla M (2006) Neues über die Verbreitung von Eragrostis albensis, E. multicaulis und E. pilosa in Österreich [New data on the distribution of Eragrostis albensis, E. multicaulis, and E. pilosa in Austria]. Linzer biol Beitr 38(2):1233–1253
Hohla M (2013) Eragrostis amurensis, Euphorbia serpens und Lepidium latifolium – neu für Oberösterreich, sowie weitere Beiträge zur Flora Österreichs. Stapfia 99:35–51
Jackowiak B, Dajdok Z, Kącki Z (2014) Corrigiola litoralis L., Nabrzeżyca nadrzeczna R Kaźmierczakowa Z Zarzycki Z Mirek Eds Polska Czerwona Księga Roślin: Paprotniki i Rośliny Kwiatowe [Polish Red Data Book of Plants: Pteridophytes and Flowering Plants], vol 3, Kraków Instytut Ochrony Przyrody, Polska Akademia Nauk, pp 131–133
Johnson LA, Soltis DE (1994) matK DNA sequence and phylogenetic reconstruction in Saxifragaceae s.s. Syst Bot 19:143–156. https://doi.org/10.2307/2419718
Kącki Z, Szczęśniak E (2009) Gatunki z rodzaju miłka Eragrostis spp [The species of the genus Eragrostis]. In: Dajdok Z, Pawlaczyk P (eds) Inwazyjne gatunki roślin ekosystemów mokradłowych Polski [Invasive plant species of wetland ecosystems in Poland]. Wydawnictwo Klubu Przyrodników [Publisher Naturalist Club], Świebodzin, pp 77–79
Király G, Virók V, Molnár VA (eds) (2011) Új magyar füvészkönyv. Magyarország hajtásos növényei [New Hungarian herb book. The vascular plants of Hungary]. Aggteleki Nemzeti Park Igazgatóság, Jósvafő
Krumbiegel A (2008) Dynamik der Uferflora in einem abschnitt der mittleren Elbe zwischen 1999 und 2007 [Dynamic of the river bank flora of a part of the middle course of the River Elbe]. Hercynia 41(1):63–82
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Lee JW, Park C-M, Rhee H (2013) Revegetation of decomposed granite roadcuts in Korea: developing digger, evaluating cost effectiveness, and determining dimensions of drilling holes, revegetation species, and mulching treatment. Land Degrad Dev 24(6):591–604. https://doi.org/10.1002/ldr.2248
Lefort V, Longueville JE, Gascuel O (2017) SMS: smart model selection in PhyML. Mol Biol Evol 34(9):2422–2424. https://doi.org/10.1093/molbev/msx149
Lomonosova, M.N. (2000) Semeystvo Zlaki—Poaceae (Gramineae). In: Krasnoborov IM (ed) Opredelitel rasteniy Novosibirskoy oblasti. Nauka—Sibirskoye predpriyatiye RAN, Novosibirsk, pp 419–451
Maillet J, Lopez-Garcia C (2000) What criteria are relevant for predicting the invasive capacity of a new agricultural weed? The case of invasive American species in France. Weed Res 40(1):11–26. https://doi.org/10.1046/j.1365-3180.2000.00171.x
Martinez B, Reaser JK, Dehgan A, Zamft B, Baisch D, McCormick C, Giordano AJ, Aicher R, Selbe S (2020) Technology innovation: advancing capacities for the early detection of and rapid response to invasive species. Biol Invasions 22:75–100. https://doi.org/10.1007/s10530-019-02146-y
Medvecká J, Kliment J, Májeková J, Halada L, Zaliberová M, Gojdičová E, Feráková V, Jarolímek I (2012) Inventory of the alien flora of Slovakia. Preslia 84:257–309
Meyer D, Buchta C (2019). proxy: distance and similarity measures. R package version 0.4–23. https://CRAN.R-project.org/package=proxy
Meyer SE, Leger EA, Eldon DR, Coleman CE (2016) Strong genetic differentiation in the invasive annual grass Bromus tectorum across the Mojave-Great Basin ecological transition zone. Biol Invasions 18:1611–1628. https://doi.org/10.1007/s10530-016-1105-6
Michalewska A, Nobis M (2005) Ekspansja Eragrostis albensis (Poaceae) na antropogenicznych siedliskach w południowo-wschodniej Polsce [Expansion of Eragrostis albensis (Poaceae) on anthropogenic sites in the south-eastern Poland]. Fragm Florist Geobot Pol 12(1):45–55
Morais P, Reichard M (2017) Cryptic invasions: a review. Sci Total Environ 613–614:1438–1448. https://doi.org/10.1016/j.scitotenv.2017.06.133
Muranaka T, Washitani I (2004) Aggressive invasion of Eragrostis curvula in gravelly floodplains of Japanese rivers: current status, ecological effects and countermeasures. Glob Environ Res 8(2):155–162
Newmaster SG, Ragupathy S (2009) Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Mol Ecol Resour 9(Suppl. 1):172–180. https://doi.org/10.1111/j.1755-0998.2009.02642.x
Nobis M, Nobis A (2009) Eragrostis pilosa (L.) P. Beauv. (Poaceae) in Poland. Biodiv Res Conserv 13:13–16
Nobis M, Nowak A, Ebel AL, Nobis A, Nowak S, Gudkova PD, Verkhozina AV, Erst AS, Łazarski G, Olonova MV, Piwowarczyk R, Bobrov AA, Khrustaleva IA, Plášek V, Silantyeva MM, Zalewska-Gałosz J (2015) Contribution to the flora of Asian and European countries: new national and regional vascular plant records, 3. Bot Lett 162(2):103–115. https://doi.org/10.1080/12538078.2015.1010105
Nylander JAA (2005) MrModeltest v2 Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden Computer program distributed by the author
Ohsako T, Ohnishi O (2000) Intra- and interspecific phylogeny of wild Fagopyrum (Polygonaceae) species based on nucleotide sequences of noncoding regions in chloroplast DNA. Am J Bot 87:573–582. https://doi.org/10.2307/2656601
Osaloo KS, Utech FH, Ohara M, Kawano S (1999) Molecular systematics of Trilliaceae I. Phylogenetic analyses of Trillium using matK gene sequences. J Plant Res 112:35–49. https://doi.org/10.1007/PL00013853
Pagitz K (2012) Eragrostis albensis neu für den Alpenraum – sowie weitere Beiträge zur Gattung Eragrostis (Eragrostideae, Poaceae) in Tirol und Österreich [Eragrostis albensis new to the Alps – and further comments on genus Eragrostis (Eragrostideae, Poaceae) for North Tyrol and Austria]. Stapfia 97:193–205
Paradis E, Schliep K (2018) ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528. https://doi.org/10.1093/bioinformatics/bty633
Parfenov VI (ed) (2013) Flora Belarusi – Sosudistye Rasteniia: Liliopsida [Flora of Belarus – Vascular Plants: Liliopsida] 2. National Academy of Sciences of Belarus, Minsk
Pejchar L, Mooney HA (2009) Invasive species, ecosystem services and human well-being. Trends Ecol Evol 24(9):497–504. https://doi.org/10.1016/j.tree.2009.03.016
Peterson PM (2003) Eragrostis Wolf. In: Barkworth ME., Capels KM., Long S, Piep MB (eds) Magnoliophyta: Commelinidae (in part): Poaceae, part 2. Flora of North America North of Mexico 25. Oxford University Press, New York, pp 65–105
Portal R (2002) Eragrostis de France et de l’Europe occidentale. Vals près Le Puy, France
Probatova NS (1985) Myatlikovye, ili Zlaki – Poaceae Bernh. (Gramineae Juss.) [Bluegrasses, or Grasses – Poaceae Bernh. (Gramineae Juss.)]. In: Charkevicz SS (ed) Plantae vasculares Orientis Extremi Sovietici 1. Lycopodiophyta, Juncaceae, Poaceae (Gramineae). Nauka, Leningrad, pp 89–382
Probatova NS, Sokolovskaya AP (1981) Chromosome numbers of some aquatic and bank plant species of the flora in the Amur River basin in connection with the peculiarities of its formation. Bot Zhur 66:1584–1594
Pyšek P, Hulme PE, Meyerson LA, Smith GF, Boatwright JS, Crouch NR, Figueiredo E, Foxcroft LC, Jarošík V, Richardson DM, Suda J and Wilson JRU (2013) Hitting the right target: taxonomic challenges for, and of, plant invasions. AoB Plants 5:plt042. https://doi.org/10.1093/aobpla/plt042
Pyšek P, Lambdon PW, Arianoutsou M, Kühn I, Pino J, Winter M (2009) Alien Vascular Plants of Europe. In: Handbook of Alien Species in Europe. Invading Nature - Springer Series in Invasion Ecology, vol 3. Springer, Dordrecht, pp 43–61. https://doi.org/10.1007/978-1-4020-8280-1_4
Pyšek P, Richardson DM (2010) Invasive species, environmental change and management, and health. Annu Rev Env Resour 35:25–55. https://doi.org/10.1146/annurev-environ-033009-095548
R Core Team (2019) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rambaut A, Suchard MA, Xie D, Drummond AJ (2014) Tracerv1.6. https://beast.bio.ed.ac.uk/Tracer
Rola K, Jędrzejczak E, Zalewska-Gałosz J, Nobis M (2019) Is the presence of simple or glandular hairs a good trait for distinguishing species in Caryophyllaceae? A case study of Arenaria serpyllifolia sensu lato in southern Poland. Acta Soc Bot Pol 88(3):3630. https://doi.org/10.5586/asbp.3630
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
RStudio Team (2016) RStudio: Integrated Development for R. RStudio, Inc., Boston, MA. http://www.rstudio.com/
Saltonstall K (2002) Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. PNAS 99(4):2445–2449. https://doi.org/10.1073/pnas.032477999
Scholz H (1995) Eragrostis albensis (Gramineae), das Elb-Liebesgras—ein neuer Neo-Endemit Mitteleuropas [Eragrostis albensis (Gramineae), the Elbe lovegrass—a new neo-endemic species of Central Europe]. Verhandlungen des Botanischen Vereins für die Provinz Brandenburg 128:73–82
Scholz H (2010) Eragrostis N.M. Wolf AP Sukhorukov (eds) The identification manual of vascular plants of the Tambov region, Tula Grif and K, pp 84–85
Scholz H, Ristow M (2005) Neue Nachrichten über die Gattung Eragrostis (Gramineae) in Mitteleuropa [New notes on the genus Eragrostis (Gramineae) in Central Europe]. Verhandlungen des Botanischen Vereins von Berlin und Brandenburg 138:15–29
Seregin AP (2012a) Taxonomic circumscription and distribution of a glandular Eurasian entity from the Eragrostis pilosa complex (Poaceae). Phytotaxa 52:8–20. https://doi.org/10.11646/phytotaxa.52.1.2
Seregin AP (2012) Floristic notes on some Eragrostis species (Gramineae) in Russia. Byul MOIP Otd biol 117(6):73–75
Shaw J, Lickey E, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166. https://doi.org/10.3732/ajb.92.1.142
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94:275–288. https://doi.org/10.3732/ajb.94.3.275
Shouliang C, Peterson PM (2006) Eragrostis Wolf. In: Zhengyi W, Raven PH, Deyuan H (eds) Flora of China 22. Science Press and Missouri Botanical Garden, Beijing and St. Louis, pp 471–479
Somaratne Y, Guan DL, Abbood NN, Zhao L, Wang WQ, Xu SQ (2019) Comparison of the complete Eragrostis pilosa chloroplast genome with its relatives in Eragrostideae (Chloridoideae; Poaceae). Plants 8(11):485. https://doi.org/10.3390/plants8110485
Špryňar P, Kubát K (2004) Eragrostis albensis a Eragrostis pectinacea, dva nové cizí druhy trav pro kvĕtenu České republiky (Poaceae) [Eragrostis albensis and Eragrostis pectinacea, two new alien grass species for the flora of the Czech Republic (Poaceae)]. Zprávy České Botanické Společnosti 39:1–24
Stöver BC, Müller KF (2010) TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform 11:7. https://doi.org/10.1186/1471-2105-11-7
Sukhorukov AP (2011) New invasive alien plant species in the forest-steppe and northern steppe subzones of European Russia: secondary range patterns, ecology and causes of fragmentary distribution. Feddes Repert 122(3–4):287–304. https://doi.org/10.1002/fedr.201100004
Swofford DL (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts
Taia WK (2006) Family Boraginaceae: hair variations and their significance in the systematic of the genera. Asian J Plant Sci 5(3):441–451. https://doi.org/10.3923/ajps.2006.441.454
Thiers B (2020, continuously updated) Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden′s Virtual Herbarium. http://sweetgum.nybg.org/ih/. Accessed on 20 Feb 2020
Tokarska-Guzik B, Dajdok Z, Zając M, Zając A, Urbisz A, Danielewicz W, Hołdyński C (2012) Rośliny obcego pochodzenia w Polsce ze szczególnym uwzględnieniem gatunków inwazyjnych [Alien plants in Poland with special reference to invasive species].The General Directorate for Environmental Protection, Warszawa
Tutin TG (1980) Eragrostis N.M.Wolf. In: Tutin TG, Heywood WH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA (eds) Alismataceae to Orchidaceae. Flora Europaea 5. Cambridge University Press, United Kingdom, pp 256–257
Tzvelev NN (1976) Zlaki SSSR [Grasses of the Soviet Union]. Nauka, Leningrad
Van den Borre A, Watson L (1994) The infrageneric classification of Eragrostis (Poaceae). Taxon 43(3):383–422. https://doi.org/10.2307/1222716
Verloove F (2010) Invaders in disguise. Conservation risks derived from misidentifications of invasive plants. Manag Biol Invasions 1(1):1–5. https://doi.org/10.3391/mbi.2010.1.1.02
Watson L, Dallwitz MJ (1992 onwards) The families of Flowering Plants: descriptions, illustrations, identification, and information retrieval. Version 2nd November 2019. https://www.delta-intkey.com/angio/www/graminea.htm. Accessed 22 Jan 2020
Wen J, Zimmer EA (1996) Phylogeny and biogeography of Panax L. (the ginseng genus, Araliaceae): Inferences from ITS sequences of nuclear ribosomal DNA. Mol Phylogenet Evol 6:167–177. https://doi.org/10.1006/mpev.1996.0069
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, London, UK, pp 315–322
Wong JXW, Costantini F, Merloni N, Savelli L, Geelen D, Airoldi L (2018) The widespread and overlooked replacement of Spartina maritima by non-indigenous S. anglica and S. townsendii in north-western Adriatic saltmarshes. Biol Invasions 20(7):1687–170. https://doi.org/10.1007/s10530-017-1654-3
Wróbel A, Nobis M (2017) Spread of Eragrostis albensis (Poaceae) and Dittrichia graveolens (Asteraceae) in the southern Poland. Acta Mus Sil Sci Nat 66:117–120. https://doi.org/10.1515/cszma-2017-0014
Wróbel A, Nobis M, Nowak A (2017) Patterns of the lemma micromorphology: a useful tool in taxonomy of the Middle Asian Eragrostis species (Poaceae). Bot Lett 164(3):253–262. https://doi.org/10.1080/23818107.2017.1339293
Yoshioka A, Kadoya T, Suda S, Washitani I (2010) Invasion of weeping lovegrass reduces native food and habitat resource of Eusphingonotus japonicus (Saussure). Biol Invasions 12(8):2789–2796. https://doi.org/10.1007/s10530-009-9684-0
Zając A, Zając M (2019) Distribution atlas of vascular plants in Poland: appendix. Institute of Botany, Jagiellonian University, with the financial support by Adam Zając & Maria Zając, Kraków
Ziętkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183. https://doi.org/10.1006/geno.1994.1151
Acknowledgements
We would like to thank the curators of the herbaria KRA, KRAM, LE, LI, WA, SZUB, and WRSL as well as the herbarium of the University of Opole for making their collections available during our research. We would like to express our appreciation to Martin Pfosser for lending us specimens labelled as E. amurensis from Upper Austria (herbarium LI), which greatly enriched our study. We are also grateful to Adam Zając (Jagiellonian University), Małgorzata Wrzesień (Maria Curie-Skłodowska University in Lublin), Zygmunt Dajdok (University of Wrocław), and Zygmunt Kącki (University of Wrocław) for sharing their data on E. albensis localities in Poland. We would like to thank two anonymous reviewers and the associate editor Carla Lambertini for their insightful and helpful feedback.
Funding
This study was partially supported by the National Science Centre, Poland, Grant Nos. 2018/29/B/NZ9/00313 and 2017/25/B/NZ8/00572.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wróbel, A., Klichowska, E., Baiakhmetov, E. et al. Invasion of Eragrostis albensis in Central Europe: distribution patterns, taxonomy and phylogenetic insight into the Eragrostis pilosa complex. Biol Invasions 23, 2305–2327 (2021). https://doi.org/10.1007/s10530-021-02507-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-021-02507-6